Role of the small subunit processome in the maintenance of pluripotent stem cells
- PMID: 26443847
- PMCID: PMC4604342
- DOI: 10.1101/gad.267112.115
Role of the small subunit processome in the maintenance of pluripotent stem cells
Abstract
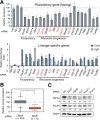
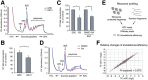
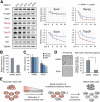
RNA-binding proteins (RBPs) play integral roles in gene regulation, yet only a small fraction of RBPs has been studied in the context of stem cells. Here we applied an RNAi screen for RBPs in mouse embryonic stem cells (ESCs) and identified 16 RBPs involved in pluripotency maintenance. Interestingly, six identified RBPs, including Krr1 and Ddx47, are part of a complex called small subunit processome (SSUP) that mediates 18S rRNA biogenesis. The SSUP components are preferentially expressed in stem cells and enhance the global translational rate, which is critical to sustain the protein levels of labile pluripotency factors such as Nanog and Esrrb. Furthermore, the SSUP proteins are required for efficient reprogramming of induced pluripotent stem cells. Our study uncovers the role of the SSUP and the importance of translational control in stem cell fate decision.
Keywords: RNA-binding protein; SSU processome; embryonic stem cell; pluripotency; ribosome; translational control.
© 2015 You et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


Similar articles
-
Regulation of pluripotency and reprogramming by RNA binding proteins.Curr Top Dev Biol. 2020;138:113-138. doi: 10.1016/bs.ctdb.2020.01.003. Epub 2020 Feb 25. Curr Top Dev Biol. 2020. PMID: 32220295 Free PMC article. Review.
-
RNA-based regulation of pluripotency.Trends Genet. 2013 Feb;29(2):99-107. doi: 10.1016/j.tig.2012.10.007. Epub 2012 Nov 10. Trends Genet. 2013. PMID: 23146412 Review.
-
MicroRNAs and RNA binding protein regulators of microRNAs in the control of pluripotency and reprogramming.Curr Opin Genet Dev. 2017 Oct;46:95-103. doi: 10.1016/j.gde.2017.07.001. Epub 2017 Jul 25. Curr Opin Genet Dev. 2017. PMID: 28753462 Review.
-
A genetic link between epigenetic repressor AS1-AS2 and a putative small subunit processome in leaf polarity establishment of Arabidopsis.Biol Open. 2016 Jul 15;5(7):942-54. doi: 10.1242/bio.019109. Biol Open. 2016. PMID: 27334696 Free PMC article.
-
Human diseases of the SSU processome.Biochim Biophys Acta. 2014 Jun;1842(6):758-64. doi: 10.1016/j.bbadis.2013.11.004. Epub 2013 Nov 12. Biochim Biophys Acta. 2014. PMID: 24240090 Free PMC article. Review.
Cited by
-
mRNA Translation Is Dynamically Regulated to Instruct Stem Cell Fate.Front Mol Biosci. 2022 Mar 31;9:863885. doi: 10.3389/fmolb.2022.863885. eCollection 2022. Front Mol Biosci. 2022. PMID: 35433828 Free PMC article. Review.
-
The Ribosomal Gene Loci-The Power behind the Throne.Genes (Basel). 2021 May 18;12(5):763. doi: 10.3390/genes12050763. Genes (Basel). 2021. PMID: 34069807 Free PMC article. Review.
-
The Best for the Most Important: Maintaining a Pristine Proteome in Stem and Progenitor Cells.Stem Cells Int. 2019 May 2;2019:1608787. doi: 10.1155/2019/1608787. eCollection 2019. Stem Cells Int. 2019. PMID: 31191665 Free PMC article. Review.
-
Response of Pluripotent Stem Cells to Environmental Stress and Its Application for Directed Differentiation.Biology (Basel). 2021 Jan 23;10(2):84. doi: 10.3390/biology10020084. Biology (Basel). 2021. PMID: 33498611 Free PMC article. Review.
-
Cardiomyogenic differentiation is fine-tuned by differential mRNA association with polysomes.BMC Genomics. 2019 Mar 15;20(1):219. doi: 10.1186/s12864-019-5550-3. BMC Genomics. 2019. PMID: 30876407 Free PMC article.
References
-
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, et al. 2010. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468: 316–320. - PubMed
-
- Deisenroth C, Zhang Y. 2010. Ribosome biogenesis surveillance: probing the ribosomal protein–Mdm2–p53 pathway. Oncogene 29: 4253–4260. - PubMed
-
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. 2009. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 4: 403–415. - PubMed
-
- Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
