Repetitive elements regulate circular RNA biogenesis
- PMID: 26442181
- PMCID: PMC4588227
- DOI: 10.1080/2159256X.2015.1045682
Repetitive elements regulate circular RNA biogenesis
Abstract
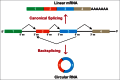
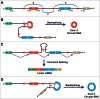
It was long assumed that eukaryotic precursor mRNAs (pre-mRNAs) are almost always spliced to generate a linear mRNA that is subsequently translated to produce a protein. However, it is now clear that thousands of protein-coding genes can be non-canonically spliced to produce circular noncoding RNAs, some of which are expressed at much higher levels than their associated linear mRNAs. How then does the splicing machinery decide whether to generate a linear mRNA or a circular RNA? Recent work has revealed that intronic repetitive elements, including sequences derived from transposons, are critical regulators of this decision. In most cases, circular RNA biogenesis appears to be initiated when complementary sequences from 2 different introns base pair to one another. This brings the splice sites from the intervening exon(s) into close proximity and facilitates the backsplicing event that generates the circular RNA. As many pre-mRNAs contain multiple intronic repeats, distinct circular transcripts can be produced depending on which repeats base pair to one another. Intronic repeats are thus critical regulatory sequences that control the functional output of their host genes, and potentially cause the functions of protein-coding genes to be highly divergent across species.
Keywords: ADAR; Alu; LINE1; backsplicing; base pairing; circRNA; circular RNA; noncoding RNA; pre-mRNA splicing; retrotransposition.
Figures

Similar articles
-
Inducible Expression of Eukaryotic Circular RNAs from Plasmids.Methods Mol Biol. 2017;1648:143-154. doi: 10.1007/978-1-4939-7204-3_11. Methods Mol Biol. 2017. PMID: 28766295
-
Circular RNAs: Unexpected outputs of many protein-coding genes.RNA Biol. 2017 Aug 3;14(8):1007-1017. doi: 10.1080/15476286.2016.1227905. Epub 2016 Aug 29. RNA Biol. 2017. PMID: 27571848 Free PMC article. Review.
-
Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins.Genes Dev. 2015 Oct 15;29(20):2168-82. doi: 10.1101/gad.270421.115. Epub 2015 Oct 8. Genes Dev. 2015. PMID: 26450910 Free PMC article.
-
A 360° view of circular RNAs: From biogenesis to functions.Wiley Interdiscip Rev RNA. 2018 Jul;9(4):e1478. doi: 10.1002/wrna.1478. Epub 2018 Apr 14. Wiley Interdiscip Rev RNA. 2018. PMID: 29655315 Free PMC article. Review.
-
Short intronic repeat sequences facilitate circular RNA production.Genes Dev. 2014 Oct 15;28(20):2233-47. doi: 10.1101/gad.251926.114. Epub 2014 Oct 3. Genes Dev. 2014. PMID: 25281217 Free PMC article.
Cited by
-
Host-derived circular RNAs display proviral activities in Hepatitis C virus-infected cells.PLoS Pathog. 2020 Aug 7;16(8):e1008346. doi: 10.1371/journal.ppat.1008346. eCollection 2020 Aug. PLoS Pathog. 2020. PMID: 32764824 Free PMC article.
-
A Multilayered Control of the Human Survival Motor Neuron Gene Expression by Alu Elements.Front Microbiol. 2017 Nov 15;8:2252. doi: 10.3389/fmicb.2017.02252. eCollection 2017. Front Microbiol. 2017. PMID: 29187847 Free PMC article. Review.
-
Circular RNAs-one of the enigmas of the brain.Neurogenetics. 2017 Jan;18(1):1-6. doi: 10.1007/s10048-016-0490-4. Epub 2016 Jul 23. Neurogenetics. 2017. PMID: 27449796 Review.
-
Human Survival Motor Neuron genes generate a vast repertoire of circular RNAs.Nucleic Acids Res. 2019 Apr 8;47(6):2884-2905. doi: 10.1093/nar/gkz034. Nucleic Acids Res. 2019. PMID: 30698797 Free PMC article.
-
Circular RNAs in Hematopoiesis with a Focus on Acute Myeloid Leukemia and Myelodysplastic Syndrome.Int J Mol Sci. 2020 Aug 19;21(17):5972. doi: 10.3390/ijms21175972. Int J Mol Sci. 2020. PMID: 32825172 Free PMC article. Review.
References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al.. Initial sequencing and analysis of the human genome. Nature 2001; 409:860-921; PMID:11237011; http://dx.doi.org/10.1038/35057062 - DOI - PubMed
-
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD.. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 2011; 7:e1002384; PMID:22144907; http://dx.doi.org/10.1371/journal.pgen.1002384 - DOI - PMC - PubMed
-
- Orgel LE, Crick FH.. Selfish DNA: the ultimate parasite. Nature 1980; 284:604-7; PMID:7366731; http://dx.doi.org/10.1038/284604a0 - DOI - PubMed
-
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV.. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 2011; 12:187-215; PMID:21801021; http://dx.doi.org/10.1146/annurev-genom-082509-141802 - DOI - PMC - PubMed
-
- Cordaux R, Batzer MA.. The impact of retrotransposons on human genome evolution. Nat Rev Genet 2009; 10:691-703; PMID:19763152; http://dx.doi.org/10.1038/nrg2640 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
