Multicopper oxidase-1 is required for iron homeostasis in Malpighian tubules of Helicoverpa armigera
- PMID: 26437857
- PMCID: PMC4593997
- DOI: 10.1038/srep14784
Multicopper oxidase-1 is required for iron homeostasis in Malpighian tubules of Helicoverpa armigera
Abstract
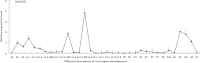
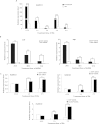
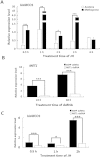
Multicopper oxidases (MCOs) are enzymes that contain 10 conserved histidine residues and 1 cysteine residue. MCO1 has been extensively investigated in the midgut because this MCO is implicated in ascorbate oxidation, iron homeostasis and immune responses. However, information regarding the action of MCO1 in Malpighian tubules is limited. In this study, Helicoverpa armigera was used as a model to investigate the function of MCO1 in Malpighian tubules. Sequence analysis results revealed that HaMCO1 exhibits typical MCO characteristics, with 10 histidine and 1 cysteine residues for copper ion binding. HaMCO1 was also found to be highly abundant in Malpighian tubules. Temporal expression patterns indicated that HaMCO1 is mainly expressed during larval molting stages. Hormone treatments [the molting hormone 20-hydroxyecdysone (20E) and juvenile hormone (JH)] revealed that 20E inhibits HaMCO1 transcript expression via its heterodimer receptor, which consists of ecdysone receptor (EcR) and ultraspiracle (USP), and that JH counteracts the action of 20E to activate HaMCO1 transcript expression via its intracellular receptor methoprene-tolerant (Met). HaMCO1 knockdown caused a significant decrease in iron accumulation and also significantly reduced transferrin and ferritin transcript expression. Therefore, HaMCO1 is coordinately regulated by 20E and JH and is required for iron homeostasis in Malpighian tubules.
Figures





Similar articles
-
Methoprene-tolerant 1 regulates gene transcription to maintain insect larval status.J Mol Endocrinol. 2014 Aug;53(1):93-104. doi: 10.1530/JME-14-0019. Epub 2014 May 28. J Mol Endocrinol. 2014. PMID: 24872508
-
Vrille is required for larval moulting and metamorphosis of Helicoverpa armigera (Lepidoptera: Noctuidae).Insect Mol Biol. 2019 Jun;28(3):355-371. doi: 10.1111/imb.12557. Epub 2019 Jan 10. Insect Mol Biol. 2019. PMID: 30485565
-
Juvenile hormone prevents 20-hydroxyecdysone-induced metamorphosis by regulating the phosphorylation of a newly identified broad protein.J Biol Chem. 2014 Sep 19;289(38):26630-26641. doi: 10.1074/jbc.M114.581876. Epub 2014 Aug 5. J Biol Chem. 2014. PMID: 25096576 Free PMC article.
-
Drosophila metamorphosis: the only way is USP?Curr Biol. 1998 Dec 3;8(24):R879-82. doi: 10.1016/s0960-9822(07)00550-7. Curr Biol. 1998. PMID: 9843674 Review.
-
Post-transcriptional regulation of insect metamorphosis and oogenesis.Cell Mol Life Sci. 2020 May;77(10):1893-1909. doi: 10.1007/s00018-019-03361-5. Epub 2019 Nov 13. Cell Mol Life Sci. 2020. PMID: 31724082 Free PMC article. Review.
Cited by
-
Ironing out the Details: Exploring the Role of Iron and Heme in Blood-Sucking Arthropods.Front Physiol. 2018 Jan 17;8:1134. doi: 10.3389/fphys.2017.01134. eCollection 2017. Front Physiol. 2018. PMID: 29387018 Free PMC article. Review.
-
Iron Homeostasis in Insects.Annu Rev Entomol. 2023 Jan 23;68:51-67. doi: 10.1146/annurev-ento-040622-092836. Epub 2022 Sep 28. Annu Rev Entomol. 2023. PMID: 36170642 Free PMC article. Review.
-
RNAi-Mediated Suppression of Laccase2 Impairs Cuticle Tanning and Molting in the Cotton Boll Weevil (Anthonomus grandis).Front Physiol. 2020 Nov 16;11:591569. doi: 10.3389/fphys.2020.591569. eCollection 2020. Front Physiol. 2020. PMID: 33329040 Free PMC article.
References
-
- Sakurai T. & Kataoka K. Basic and applied features of multicopper oxidases, CueO, bilirubin oxidase, and laccase. Chem. Rec. 7, 220–229 (2007). - PubMed
-
- Dittmer N. T. & Kanost M. R. Insect multicopper oxidases: diversity, properties, and physiological roles. Insect Biochem. Mol. Biol. 40, 179–188 (2010). - PubMed
-
- Hoegger P. J., Kilaru S., James T. Y., Thacker J. R. & Kües U. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 273, 2308–2326 (2006). - PubMed
-
- Dittmer N. T. et al. Characterization of cDNAs encoding putative laccase-like multicopper oxidases and developmental expression in the tobacco hornworm, Manduca sexta, and the malaria mosquito, Anopheles gambiae. Insect Biochem. Mol. Biol. 34, 29–41 (2004). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

