New Insights into the Biological Role of Mammalian ADARs; the RNA Editing Proteins
- PMID: 26437436
- PMCID: PMC4693238
- DOI: 10.3390/biom5042338
New Insights into the Biological Role of Mammalian ADARs; the RNA Editing Proteins
Abstract
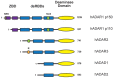
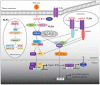
The ADAR proteins deaminate adenosine to inosine in double-stranded RNA which is one of the most abundant modifications present in mammalian RNA. Inosine can have a profound effect on the RNAs that are edited, not only changing the base-pairing properties, but can also result in recoding, as inosine behaves as if it were guanosine. In mammals there are three ADAR proteins and two ADAR-related proteins (ADAD) expressed. All have a very similar modular structure; however, both their expression and biological function differ significantly. Only two of the ADAR proteins have enzymatic activity. However, both ADAR and ADAD proteins possess the ability to bind double-strand RNA. Mutations in ADARs have been associated with many diseases ranging from cancer, innate immunity to neurological disorders. Here, we will discuss in detail the domain structure of mammalian ADARs, the effects of RNA editing, and the role of ADARs in human diseases.
Keywords: ADAR; Alu elements; RNA editing; cancer; deaminase domain; dsRBDs.
Figures
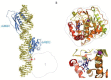
Similar articles
-
RNA editing by mammalian ADARs.Adv Genet. 2011;73:87-120. doi: 10.1016/B978-0-12-380860-8.00003-3. Adv Genet. 2011. PMID: 21310295 Review.
-
Mechanisms and implications of ADAR-mediated RNA editing in cancer.Cancer Lett. 2017 Dec 28;411:27-34. doi: 10.1016/j.canlet.2017.09.036. Epub 2017 Sep 30. Cancer Lett. 2017. PMID: 28974449 Review.
-
To protect and modify double-stranded RNA - the critical roles of ADARs in development, immunity and oncogenesis.Crit Rev Biochem Mol Biol. 2021 Feb;56(1):54-87. doi: 10.1080/10409238.2020.1856768. Epub 2020 Dec 27. Crit Rev Biochem Mol Biol. 2021. PMID: 33356612 Free PMC article. Review.
-
How do ADARs bind RNA? New protein-RNA structures illuminate substrate recognition by the RNA editing ADARs.Bioessays. 2017 Apr;39(4):10.1002/bies.201600187. doi: 10.1002/bies.201600187. Epub 2017 Feb 20. Bioessays. 2017. PMID: 28217931 Free PMC article. Review.
-
In cancer, A-to-I RNA editing can be the driver, the passenger, or the mechanic.Drug Resist Updat. 2017 May;32:16-22. doi: 10.1016/j.drup.2017.09.001. Epub 2017 Oct 4. Drug Resist Updat. 2017. PMID: 29145975 Review.
Cited by
-
Sensing of transposable elements by the antiviral innate immune system.RNA. 2021 Apr 22;27(7):735-52. doi: 10.1261/rna.078721.121. Online ahead of print. RNA. 2021. PMID: 33888553 Free PMC article.
-
Dynamic temperature-sensitive A-to-I RNA editing in the brain of a heterothermic mammal during hibernation.RNA. 2018 Nov;24(11):1481-1495. doi: 10.1261/rna.066522.118. Epub 2018 Jul 31. RNA. 2018. PMID: 30065024 Free PMC article.
-
Post-transcriptional regulation of LINE-1 retrotransposition by AID/APOBEC and ADAR deaminases.Chromosome Res. 2018 Mar;26(1-2):45-59. doi: 10.1007/s10577-018-9572-5. Epub 2018 Feb 2. Chromosome Res. 2018. PMID: 29396793 Review.
-
The Epitranscriptome and Innate Immunity.PLoS Genet. 2015 Dec 10;11(12):e1005687. doi: 10.1371/journal.pgen.1005687. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26658668 Free PMC article. Review.
-
Natural Antisense Transcripts at the Interface between Host Genome and Mobile Genetic Elements.Front Microbiol. 2017 Nov 20;8:2292. doi: 10.3389/fmicb.2017.02292. eCollection 2017. Front Microbiol. 2017. PMID: 29209299 Free PMC article.
References
-
- Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M., et al. Modomics: A database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

