Transgenic Cotton Plants Expressing Double-stranded RNAs Target HMG-CoA Reductase (HMGR) Gene Inhibits the Growth, Development and Survival of Cotton Bollworms
- PMID: 26435695
- PMCID: PMC4582153
- DOI: 10.7150/ijbs.12463
Transgenic Cotton Plants Expressing Double-stranded RNAs Target HMG-CoA Reductase (HMGR) Gene Inhibits the Growth, Development and Survival of Cotton Bollworms
Abstract
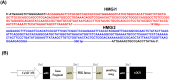
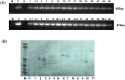
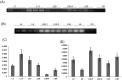
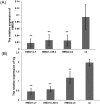
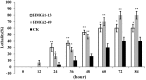
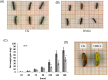
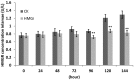
RNA interference (RNAi) has been developed as a powerful technique in the research of functional genomics as well as plant pest control. In this report, double-stranded RNAs (dsRNA) targeting 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) gene, which catalyze a rate-limiting enzymatic reaction in the mevalonate pathway of juvenile hormone (JH) synthesis in cotton bollworm, was expressed in cotton plants via Agrobacterium tumefaciens-mediated transformation. PCR and Sothern analysis revealed the integration of HMGR gene into cotton genome. RT-PCR and qRT-PCR confirmed the high transcription level of dsHMGR in transgenic cotton lines. The HMGR expression both in transcription and translation level was significantly downregulated in cotton bollworms (helicoverpa armigera) larvae after feeding on the leaves of HMGR transgenic plants. The transcription level of HMGR gene in larvae reared on transgenic cotton leaves was as much as 80.68% lower than that of wild type. In addition, the relative expression level of vitellogenin (Vg, crucial source of nourishment for offspring embryo development) gene was also reduced by 76.86% when the insect larvae were fed with transgenic leaves. The result of insect bioassays showed that the transgenic plant harboring dsHMGR not only inhibited net weight gain but also delayed the growth of cotton bollworm larvae. Taken together, transgenic cotton plant expressing dsRNAs successfully downregulated HMGR gene and impaired the development and survival of target insect, which provided more option for plant pest control.
Keywords: 3-hydroxy-3-methylglutaryl coenzyme A reductase(HMGR); RNA interference; cotton bollworm; double-stranded RNAs; pest control.; transgenic cotton.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

Similar articles
-
Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms.Transgenic Res. 2011 Jun;20(3):665-73. doi: 10.1007/s11248-010-9450-1. Epub 2010 Oct 17. Transgenic Res. 2011. PMID: 20953975 Free PMC article.
-
RNAi silencing of the HaHMG-CoA reductase gene inhibits oviposition in the Helicoverpa armigera cotton bollworm.PLoS One. 2013 Jul 2;8(7):e67732. doi: 10.1371/journal.pone.0067732. Print 2013. PLoS One. 2013. PMID: 23844078 Free PMC article.
-
Transgenic Cotton Plants Expressing the HaHR3 Gene Conferred Enhanced Resistance to Helicoverpa armigera and Improved Cotton Yield.Int J Mol Sci. 2017 Aug 30;18(9):1874. doi: 10.3390/ijms18091874. Int J Mol Sci. 2017. PMID: 28867769 Free PMC article.
-
Early detection of field-evolved resistance to Bt cotton in China: cotton bollworm and pink bollworm.J Invertebr Pathol. 2012 Jul;110(3):301-6. doi: 10.1016/j.jip.2012.04.008. Epub 2012 Apr 16. J Invertebr Pathol. 2012. PMID: 22537835 Review.
-
Diet-delivered RNAi in Helicoverpa armigera--Progresses and challenges.J Insect Physiol. 2016 Feb;85:86-93. doi: 10.1016/j.jinsphys.2015.11.005. Epub 2015 Nov 5. J Insect Physiol. 2016. PMID: 26549127 Review.
Cited by
-
RNA interference as a gene silencing tool to control Tuta absoluta in tomato (Solanum lycopersicum).PeerJ. 2016 Dec 15;4:e2673. doi: 10.7717/peerj.2673. eCollection 2016. PeerJ. 2016. PMID: 27994959 Free PMC article.
-
High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system.Plant Biotechnol J. 2018 Jan;16(1):137-150. doi: 10.1111/pbi.12755. Epub 2017 Jun 20. Plant Biotechnol J. 2018. PMID: 28499063 Free PMC article.
-
Biosynthetic pathways of triterpenoids and strategies to improve their Biosynthetic Efficiency.Plant Growth Regul. 2022;97(3):439-454. doi: 10.1007/s10725-022-00818-9. Epub 2022 Mar 31. Plant Growth Regul. 2022. PMID: 35382096 Free PMC article. Review.
-
Host-Delivered RNA Interference for Durable Pest Resistance in Plants: Advanced Methods, Challenges, and Applications.Mol Biotechnol. 2024 Aug;66(8):1786-1805. doi: 10.1007/s12033-023-00833-9. Epub 2023 Jul 31. Mol Biotechnol. 2024. PMID: 37523020 Review.
-
Overexpression of soybean trypsin inhibitor genes decreases defoliation by corn earworm (Helicoverpa zea) in soybean (Glycine max) and Arabidopsis thaliana.Front Plant Sci. 2023 Feb 17;14:1129454. doi: 10.3389/fpls.2023.1129454. eCollection 2023. Front Plant Sci. 2023. PMID: 36875574 Free PMC article.
References
-
- Wu KM, Lu YH, Feng HQ, Jiang YY, Zhao JZ. Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science. 2008;321:1676–8. - PubMed
-
- Bravo A, Soberon M. How to cope with insect resistance to Bt toxins? Trends in biotechnology. 2008;26:573–9. - PubMed
-
- Tabashnik BE, Gassmann AJ, Crowder DW, Carriere Y. Insect resistance to Bt crops: evidence versus theory. Nature biotechnology. 2008;26:199–202. - PubMed
-
- Tabashnik BE, Brevault T, Carriere Y. Insect resistance to Bt crops: lessons from the first billion acres. Nat Biotechnol. 2013;31:510–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

