Novel Role of TRPML2 in the Regulation of the Innate Immune Response
- PMID: 26432893
- PMCID: PMC4637233
- DOI: 10.4049/jimmunol.1500163
Novel Role of TRPML2 in the Regulation of the Innate Immune Response
Abstract
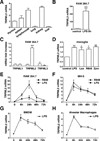
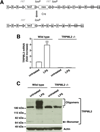
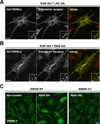
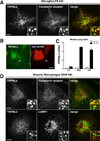
TRPMLs (or mucolipins) constitute a family of endosomal cation channels with homology to the transient receptor potential superfamily. In mammals, the TRPML family includes three members: TRPML1-3. Although TRPML1 and TRPML3 have been well characterized, the cellular function of TRPML2 has remained elusive. To address TRPML2 function in a physiologically relevant cell type, we first analyzed TRPML2 expression in different mouse tissues and organs and found that it was predominantly expressed in lymphoid organs and kidney. Quantitative RT-PCR revealed tight regulation of TRPML2 at the transcriptional level. Although TRPML2 expression was negligible in resting macrophages, TRPML2 mRNA and protein levels dramatically increased in response to TLR activation both in vitro and in vivo. Conversely, TRPML1 and TRPML3 levels did not change upon TLR activation. Immunofluorescence analysis demonstrated that endogenous TRPML2 primarily localized to recycling endosomes both in culture and primary cells, in contrast with TRPML1 and TRPML3, which distribute to the late and early endosomal pathway, respectively. To better understand the in vivo function of TRPML2, we generated a TRPML2-knockout mouse. We found that the production of several chemokines, in particular CCL2, was severely reduced in TRPML2-knockout mice. Furthermore, TRPML2-knockout mice displayed impaired recruitment of peripheral macrophages in response to i.p. injections of LPS or live bacteria, suggesting a potential defect in the immune response. Overall, our study reveals interesting differences in the regulation and distribution of the members of the TRPML family and identifies a novel role for TRPML2 in the innate immune response.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Gene expression of TRPMLs and its regulation by pathogen stimulation.Gene. 2023 May 15;864:147291. doi: 10.1016/j.gene.2023.147291. Epub 2023 Feb 20. Gene. 2023. PMID: 36813061
-
TRPML2 and mucolipin evolution.Handb Exp Pharmacol. 2014;222:647-58. doi: 10.1007/978-3-642-54215-2_25. Handb Exp Pharmacol. 2014. PMID: 24756724 Review.
-
Involvement of the TRPML Mucolipin Channels in Viral Infections and Anti-viral Innate Immune Responses.Front Immunol. 2020 Apr 29;11:739. doi: 10.3389/fimmu.2020.00739. eCollection 2020. Front Immunol. 2020. PMID: 32425938 Free PMC article. Review.
-
The tissue-specific expression of TRPML2 (MCOLN-2) gene is influenced by the presence of TRPML1.Pflugers Arch. 2009 Nov;459(1):79-91. doi: 10.1007/s00424-009-0716-5. Pflugers Arch. 2009. PMID: 19763610 Free PMC article.
-
Lysosomal localization of TRPML3 depends on TRPML2 and the mucolipidosis-associated protein TRPML1.J Biol Chem. 2006 Jun 23;281(25):17517-17527. doi: 10.1074/jbc.M600807200. Epub 2006 Apr 10. J Biol Chem. 2006. PMID: 16606612 Free PMC article.
Cited by
-
The role of stretch-activated ion channels in acute respiratory distress syndrome: finally a new target?Am J Physiol Lung Cell Mol Physiol. 2016 Sep 1;311(3):L639-52. doi: 10.1152/ajplung.00458.2015. Epub 2016 Aug 12. Am J Physiol Lung Cell Mol Physiol. 2016. PMID: 27521425 Free PMC article. Review.
-
The Different Immune Responses by Age Are due to the Ability of the Fetal Immune System to Secrete Primal Immunoglobulins Responding to Unexperienced Antigens.Int J Biol Sci. 2022 Jan 1;18(2):617-636. doi: 10.7150/ijbs.67203. eCollection 2022. Int J Biol Sci. 2022. PMID: 35002513 Free PMC article.
-
Function of TRP channels in monocytes/macrophages.Front Immunol. 2023 Jun 19;14:1187890. doi: 10.3389/fimmu.2023.1187890. eCollection 2023. Front Immunol. 2023. PMID: 37404813 Free PMC article. Review.
-
The role of lysosomes in cancer development and progression.Cell Biosci. 2020 Nov 18;10(1):131. doi: 10.1186/s13578-020-00489-x. Cell Biosci. 2020. PMID: 33292489 Free PMC article. Review.
-
Pathological Functions of Lysosomal Ion Channels in the Central Nervous System.Int J Mol Sci. 2024 Jun 14;25(12):6565. doi: 10.3390/ijms25126565. Int J Mol Sci. 2024. PMID: 38928271 Free PMC article. Review.
References
-
- Bargal R, Avidan N, Ben-Asher E, Olender Z, Zeigler M, Frumkin A, Raas-Rothschild A, Glusman G, Lancet D, Bach G. Identification of the gene causing mucolipidosis type IV. Nature genetics. 2000;26:118–123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

