Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine
- PMID: 26424849
- PMCID: PMC4678861
- DOI: 10.1093/nar/gkv980
Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine
Abstract
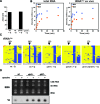
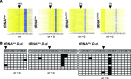
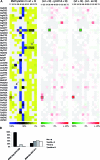
Dnmt2 enzymes are cytosine-5 methyltransferases that methylate C38 of several tRNAs. We report here that the activities of two Dnmt2 homologs, Pmt1 from Schizosaccharomyces pombe and DnmA from Dictyostelium discoideum, are strongly stimulated by prior queuosine (Q) modification of the substrate tRNA. In vivo tRNA methylation levels were stimulated by growth of cells in queuine-containing medium; in vitro Pmt1 activity was enhanced on Q-containing RNA; and queuine-stimulated in vivo methylation was abrogated by the absence of the enzyme that inserts queuine into tRNA, eukaryotic tRNA-guanine transglycosylase. Global analysis of tRNA methylation in S. pombe showed a striking selectivity of Pmt1 for tRNA(Asp) methylation, which distinguishes Pmt1 from other Dnmt2 homologs. The present analysis also revealed a novel Pmt1- and Q-independent tRNA methylation site in S. pombe, C34 of tRNA(Pro). Notably, queuine is a micronutrient that is scavenged by higher eukaryotes from the diet and gut microflora. This work therefore reveals an unanticipated route by which the environment can modulate tRNA modification in an organism.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Cross-Talk between Dnmt2-Dependent tRNA Methylation and Queuosine Modification.Biomolecules. 2017 Feb 10;7(1):14. doi: 10.3390/biom7010014. Biomolecules. 2017. PMID: 28208632 Free PMC article. Review.
-
Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling.Nucleic Acids Res. 2012 Dec;40(22):11648-58. doi: 10.1093/nar/gks956. Epub 2012 Oct 15. Nucleic Acids Res. 2012. PMID: 23074192 Free PMC article.
-
Structural insights into the stimulation of S. pombe Dnmt2 catalytic efficiency by the tRNA nucleoside queuosine.Sci Rep. 2018 Jun 11;8(1):8880. doi: 10.1038/s41598-018-27118-5. Sci Rep. 2018. PMID: 29892076 Free PMC article.
-
Queuine links translational control in eukaryotes to a micronutrient from bacteria.Nucleic Acids Res. 2019 Apr 23;47(7):3711-3727. doi: 10.1093/nar/gkz063. Nucleic Acids Res. 2019. PMID: 30715423 Free PMC article.
-
Queuosine modification of tRNA: its divergent role in cellular machinery.Biosci Rep. 2009 Nov 23;30(2):135-48. doi: 10.1042/BSR20090057. Biosci Rep. 2009. PMID: 19925456 Review.
Cited by
-
Origins and evolving functionalities of tRNA-derived small RNAs.Trends Biochem Sci. 2021 Oct;46(10):790-804. doi: 10.1016/j.tibs.2021.05.001. Epub 2021 May 27. Trends Biochem Sci. 2021. PMID: 34053843 Free PMC article. Review.
-
Position 34 of tRNA is a discriminative element for m5C38 modification by human DNMT2.Nucleic Acids Res. 2021 Dec 16;49(22):13045-13061. doi: 10.1093/nar/gkab1148. Nucleic Acids Res. 2021. PMID: 34871455 Free PMC article.
-
To be or not to be modified: Miscellaneous aspects influencing nucleotide modifications in tRNAs.IUBMB Life. 2019 Aug;71(8):1126-1140. doi: 10.1002/iub.2041. Epub 2019 Apr 1. IUBMB Life. 2019. PMID: 30932315 Free PMC article. Review.
-
Dynamic queuosine changes in tRNA couple nutrient levels to codon choice in Trypanosoma brucei.Nucleic Acids Res. 2021 Dec 16;49(22):12986-12999. doi: 10.1093/nar/gkab1204. Nucleic Acids Res. 2021. PMID: 34883512 Free PMC article.
-
Different modification pathways for m1A58 incorporation in yeast elongator and initiator tRNAs.Nucleic Acids Res. 2023 Oct 27;51(19):10653-10667. doi: 10.1093/nar/gkad722. Nucleic Acids Res. 2023. PMID: 37650648 Free PMC article.
References
-
- Motorin Y., Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. - PubMed
-
- El Yacoubi B., Bailly M., de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012;46:69–95. - PubMed
-
- Motorin Y., Helm M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA. 2011;2:611–631. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

