Characterization of structural elements in native autoinducing peptides and non-native analogues that permit the differential modulation of AgrC-type quorum sensing receptors in Staphylococcus aureus
- PMID: 26416476
- PMCID: PMC4681674
- DOI: 10.1039/c5ob01735a
Characterization of structural elements in native autoinducing peptides and non-native analogues that permit the differential modulation of AgrC-type quorum sensing receptors in Staphylococcus aureus
Abstract
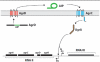
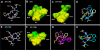
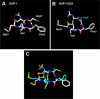
Staphylococcus aureus uses short macrocyclic peptides (i.e., autoinducing peptides, or AIPs) to assess its local population density in a cell-cell signaling mechanism called quorum sensing (QS). At high cell numbers, this pathogen can initiate many virulent behaviors that allow for the establishment of infection. Binding of the AIP signal to its cognate transmembrane AgrC-type receptor is a critical event in the QS signaling cascade; consequently, interference of AIP:receptor interactions may have the potential to prevent and eradicate certain S. aureus infections. To date, four pairs of AIP:AgrC receptors have been identified in S. aureus, each pair being utilized by a specific S. aureus group (I-IV). Other staphylococcal species also use closely related, but distinct, AIP:AgrC pairs to control QS. We seek to develop non-native ligands capable of intercepting AIP:AgrC binding in each S. aureus group and in related species. As these bacteria may use their respective AIP signal to attenuate the QS systems of other groups/species, such ligands would provide valuable chemical tools to probe possible interference mechanisms in a range of contexts. In the current study, we used solution-phase NMR techniques to characterize the 3-D structures of a set of known native and non-native peptides that have differential modulatory activity in certain AgrC receptors. Analysis of these structures revealed several distinct structural motifs that belay differential activity in selected S. aureus AgrC receptors (i.e., AgrC-I, AgrC-II, and AgrC-III). The results of this study can be leveraged for the design of new synthetic ligands with enhanced selectivities and potencies for these AgrC receptors.
Figures
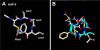

Similar articles
-
Simplified Autoinducing Peptide Mimetics with Single-Nanomolar Activity Against the Staphylococcus aureus AgrC Quorum Sensing Receptor.ACS Infect Dis. 2019 Apr 12;5(4):484-492. doi: 10.1021/acsinfecdis.9b00002. Epub 2019 Mar 13. ACS Infect Dis. 2019. PMID: 30817121 Free PMC article.
-
Structural characterization of native autoinducing peptides and abiotic analogues reveals key features essential for activation and inhibition of an AgrC quorum sensing receptor in Staphylococcus aureus.J Am Chem Soc. 2013 Dec 11;135(49):18436-44. doi: 10.1021/ja407533e. Epub 2013 Nov 25. J Am Chem Soc. 2013. PMID: 24219181 Free PMC article.
-
Highly potent inhibitors of quorum sensing in Staphylococcus aureus revealed through a systematic synthetic study of the group-III autoinducing peptide.J Am Chem Soc. 2013 May 29;135(21):7869-82. doi: 10.1021/ja3112115. Epub 2013 May 17. J Am Chem Soc. 2013. PMID: 23647400
-
Quorum-sensing, intra- and inter-species competition in the staphylococci.Microbiology (Reading). 2023 Aug;169(8):001381. doi: 10.1099/mic.0.001381. Microbiology (Reading). 2023. PMID: 37578829 Free PMC article. Review.
-
Quorum Quenching Strategy Targeting Gram-Positive Pathogenic Bacteria.Adv Exp Med Biol. 2016;901:109-30. doi: 10.1007/5584_2016_1. Adv Exp Med Biol. 2016. PMID: 27167409 Review.
Cited by
-
Regulation of Virulence in Staphylococcus aureus: Molecular Mechanisms and Remaining Puzzles.Cell Chem Biol. 2016 Feb 18;23(2):214-224. doi: 10.1016/j.chembiol.2016.01.004. Cell Chem Biol. 2016. PMID: 26971873 Free PMC article. Review.
-
Non-Native Peptides Capable of Pan-Activating the agr Quorum Sensing System across Multiple Specificity Groups of Staphylococcus epidermidis.ACS Chem Biol. 2021 Jun 18;16(6):1070-1078. doi: 10.1021/acschembio.1c00240. Epub 2021 May 14. ACS Chem Biol. 2021. PMID: 33988969 Free PMC article.
-
Characterization of an Autoinducing Peptide Signal Reveals Highly Efficacious Synthetic Inhibitors and Activators of Quorum Sensing and Biofilm Formation in Listeria monocytogenes.Biochemistry. 2023 Oct 3;62(19):2878-2892. doi: 10.1021/acs.biochem.3c00373. Epub 2023 Sep 12. Biochemistry. 2023. PMID: 37699554 Free PMC article.
-
Simplified Autoinducing Peptide Mimetics with Single-Nanomolar Activity Against the Staphylococcus aureus AgrC Quorum Sensing Receptor.ACS Infect Dis. 2019 Apr 12;5(4):484-492. doi: 10.1021/acsinfecdis.9b00002. Epub 2019 Mar 13. ACS Infect Dis. 2019. PMID: 30817121 Free PMC article.
-
Conformational Switch to a β-Turn in a Staphylococcal Quorum Sensing Signal Peptide Causes a Dramatic Increase in Potency.J Am Chem Soc. 2020 Jan 15;142(2):750-761. doi: 10.1021/jacs.9b05513. Epub 2020 Jan 2. J Am Chem Soc. 2020. PMID: 31859506 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

