Mechanism of MenE inhibition by acyl-adenylate analogues and discovery of novel antibacterial agents
- PMID: 26394156
- PMCID: PMC4624480
- DOI: 10.1021/acs.biochem.5b00966
Mechanism of MenE inhibition by acyl-adenylate analogues and discovery of novel antibacterial agents
Abstract
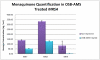
MenE is an o-succinylbenzoyl-CoA (OSB-CoA) synthetase in the bacterial menaquinone biosynthesis pathway and is a promising target for the development of novel antibacterial agents. The enzyme catalyzes CoA ligation via an acyl-adenylate intermediate, and we have previously reported tight-binding inhibitors of MenE based on stable acyl-sulfonyladenosine analogues of this intermediate, including OSB-AMS (1), which has an IC50 value of ≤25 nM for Escherichia coli MenE. Herein, we show that OSB-AMS reduces menaquinone levels in Staphylococcus aureus, consistent with its proposed mechanism of action, despite the observation that the antibacterial activity of OSB-AMS is ∼1000-fold lower than the IC50 for enzyme inhibition. To inform the synthesis of MenE inhibitors with improved antibacterial activity, we have undertaken a structure-activity relationship (SAR) study stimulated by the knowledge that OSB-AMS can adopt two isomeric forms in which the OSB side chain exists either as an open-chain keto acid or a cyclic lactol. These studies revealed that negatively charged analogues of the keto acid form bind, while neutral analogues do not, consistent with the hypothesis that the negatively charged keto acid form of OSB-AMS is the active isomer. X-ray crystallography and site-directed mutagenesis confirm the importance of a conserved arginine for binding the OSB carboxylate. Although most lactol isomers tested were inactive, a novel difluoroindanediol inhibitor (11) with improved antibacterial activity was discovered, providing a pathway toward the development of optimized MenE inhibitors in the future.
Figures


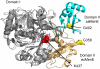
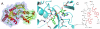
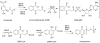
Similar articles
-
Structure-Based Design, Synthesis, and Biological Evaluation of Non-Acyl Sulfamate Inhibitors of the Adenylate-Forming Enzyme MenE.Biochemistry. 2019 Apr 9;58(14):1918-1930. doi: 10.1021/acs.biochem.9b00003. Epub 2019 Mar 26. Biochemistry. 2019. PMID: 30912442 Free PMC article.
-
Stable analogues of OSB-AMP: potent inhibitors of MenE, the o-succinylbenzoate-CoA synthetase from bacterial menaquinone biosynthesis.Chembiochem. 2012 Jan 2;13(1):129-36. doi: 10.1002/cbic.201100585. Epub 2011 Nov 23. Chembiochem. 2012. PMID: 22109989 Free PMC article.
-
Crystal structure of the thioesterification conformation of Bacillus subtilis o-succinylbenzoyl-CoA synthetase reveals a distinct substrate-binding mode.J Biol Chem. 2017 Jul 21;292(29):12296-12310. doi: 10.1074/jbc.M117.790410. Epub 2017 May 30. J Biol Chem. 2017. PMID: 28559280 Free PMC article.
-
Structure guided design of biotin protein ligase inhibitors for antibiotic discovery.Curr Top Med Chem. 2014;14(1):4-20. doi: 10.2174/1568026613666131111103149. Curr Top Med Chem. 2014. PMID: 24236729 Review.
-
DD-ligases as a potential target for antibiotics: past, present and future.Curr Med Chem. 2009;16(20):2566-80. doi: 10.2174/092986709788682029. Curr Med Chem. 2009. PMID: 19601798 Review.
Cited by
-
Structure-Based Design, Synthesis, and Biological Evaluation of Non-Acyl Sulfamate Inhibitors of the Adenylate-Forming Enzyme MenE.Biochemistry. 2019 Apr 9;58(14):1918-1930. doi: 10.1021/acs.biochem.9b00003. Epub 2019 Mar 26. Biochemistry. 2019. PMID: 30912442 Free PMC article.
-
Structural approaches to pathway-specific antimicrobial agents.Transl Res. 2020 Jun;220:114-121. doi: 10.1016/j.trsl.2020.02.001. Epub 2020 Feb 6. Transl Res. 2020. PMID: 32105648 Free PMC article. Review.
-
Novel long-chain compounds with both immunomodulatory and MenA inhibitory activities against Staphylococcus aureus and its biofilm.Sci Rep. 2017 Jan 10;7:40077. doi: 10.1038/srep40077. Sci Rep. 2017. PMID: 28071679 Free PMC article.
-
A Novel Small-Molecule Inhibitor of the Mycobacterium tuberculosis Demethylmenaquinone Methyltransferase MenG Is Bactericidal to Both Growing and Nutritionally Deprived Persister Cells.mBio. 2017 Feb 14;8(1):e02022-16. doi: 10.1128/mBio.02022-16. mBio. 2017. PMID: 28196957 Free PMC article.
-
Targeting adenylate-forming enzymes with designed sulfonyladenosine inhibitors.J Antibiot (Tokyo). 2019 Jun;72(6):325-349. doi: 10.1038/s41429-019-0171-2. Epub 2019 Apr 15. J Antibiot (Tokyo). 2019. PMID: 30982830 Free PMC article. Review.
References
-
- Simmons KJ, Chopra I, Fishwick CW. Structure-based discovery of antibacterial drugs. Nat Rev Microbiol. 2010;8(7):501–510. - PubMed
-
- Paul S, et al. New and emerging pathogens. Part 5. Those crazy cocci: more virulent and resistant to antibiotics than ever. MLO Med Lab Obs. 1996;28(6):40–42. 44. 46 passim. - PubMed
-
- Harper C. Tuberculosis, a neglected opportunity? Nat Med. 2007;13(3):309–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Medical
Molecular Biology Databases
Miscellaneous

