COP9 signalosome controls the degradation of cytosolic misfolded proteins and protects against cardiac proteotoxicity
- PMID: 26383969
- PMCID: PMC4636927
- DOI: 10.1161/CIRCRESAHA.115.306783
COP9 signalosome controls the degradation of cytosolic misfolded proteins and protects against cardiac proteotoxicity
Abstract
Rationale: Impaired degradation of misfolded proteins is associated with a large subset of heart diseases. Misfolded proteins are degraded primarily by the ubiquitin-proteasome system, but the ubiquitin ligases responsible for the degradation remain largely unidentified. The cullin deneddylation activity of the COP9 signalosome (CSN) requires all 8 CSN subunits (CSN1 through CSN8) and regulates cullin-RING ligases, thereby controlling ubiquitination of a large number of proteins; however, neither CSN nor cullin-RING ligases is known to regulate the degradation of cytosolic misfolded proteins.
Objective: We sought to investigate the role of CSN8/CSN in misfolded protein degradation and cardiac proteinopathy.
Methods and results: Cardiac CSN8 knockout causes mouse premature death; hence, CSN8 hypomorphism (CSN8(hypo)) mice were used. Myocardial neddylated forms of cullins were markedly increased, and myocardial capacity of degrading a surrogate misfolded protein was significantly reduced by CSN8 hypomorphism. When introduced into proteinopathic mice in which a bona fide misfolded protein R120G missense mutation of αβ-crystallin (CryAB(R120G)) is overexpressed in the heart, CSN8 hypomorphism aggravated CryAB(R120G)-induced restrictive cardiomyopathy and shortened the lifespan of CryAB(R120G) mice, which was associated with augmented accumulation of protein aggregates, increased neddylated proteins, and reduced levels of total ubiquitinated proteins and LC3-II in the heart. In cultured cardiomyocytes, both CSN8 knockdown and cullin-RING ligase inactivation suppressed the ubiquitination and degradation of CryAB(R120G) but not native CryAB, resulting in accumulation of protein aggregates and exacerbation of CryAB(R120G) cytotoxicity.
Conclusions: (1) CSN8/CSN promotes the ubiquitination and degradation of misfolded proteins and protects against cardiac proteotoxicity, and (2) cullin-RING ligases participate in degradation of cytosolic misfolded proteins.
Keywords: COP9 signalosome; Cops8; autophagy; desmin-related cardiomyopathy; proteasome; ubiquitin.
© 2015 American Heart Association, Inc.
Conflict of interest statement
The authors declare there is no conflict of interest to disclose.
Figures

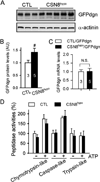
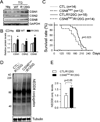
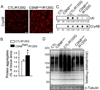
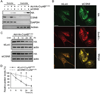

Comment in
-
Breaking down the COP9 Signalsome in the heart: how inactivating a protein ubiquitin ligase increases protein ubiquitylation and protects the heart.Circ Res. 2015 Nov 6;117(11):914-6. doi: 10.1161/CIRCRESAHA.115.307644. Circ Res. 2015. PMID: 26541679 Free PMC article. No abstract available.
Similar articles
-
The COP9 signalosome is required for autophagy, proteasome-mediated proteolysis, and cardiomyocyte survival in adult mice.Circ Heart Fail. 2013 Sep 1;6(5):1049-57. doi: 10.1161/CIRCHEARTFAILURE.113.000338. Epub 2013 Jul 19. Circ Heart Fail. 2013. PMID: 23873473 Free PMC article.
-
Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice.Circ Res. 2011 Jan 7;108(1):40-50. doi: 10.1161/CIRCRESAHA.110.230607. Epub 2010 Nov 4. Circ Res. 2011. PMID: 21051661 Free PMC article.
-
The COP9 signalosome coerces autophagy and the ubiquitin-proteasome system to police the heart.Autophagy. 2016;12(3):601-2. doi: 10.1080/15548627.2015.1136773. Autophagy. 2016. PMID: 26760900 Free PMC article.
-
Roles of COP9 signalosome in cancer.Cell Cycle. 2011 Sep 15;10(18):3057-66. doi: 10.4161/cc.10.18.17320. Epub 2011 Sep 15. Cell Cycle. 2011. PMID: 21876386 Free PMC article. Review.
-
Deregulation of the COP9 signalosome-cullin-RING ubiquitin-ligase pathway: mechanisms and roles in urological cancers.Int J Biochem Cell Biol. 2013 Jul;45(7):1327-37. doi: 10.1016/j.biocel.2013.03.023. Epub 2013 Apr 10. Int J Biochem Cell Biol. 2013. PMID: 23583660 Review.
Cited by
-
Breaking down the COP9 Signalsome in the heart: how inactivating a protein ubiquitin ligase increases protein ubiquitylation and protects the heart.Circ Res. 2015 Nov 6;117(11):914-6. doi: 10.1161/CIRCRESAHA.115.307644. Circ Res. 2015. PMID: 26541679 Free PMC article. No abstract available.
-
Neddylation mediates ventricular chamber maturation through repression of Hippo signaling.Proc Natl Acad Sci U S A. 2018 Apr 24;115(17):E4101-E4110. doi: 10.1073/pnas.1719309115. Epub 2018 Apr 9. Proc Natl Acad Sci U S A. 2018. PMID: 29632206 Free PMC article.
-
Systemic inhibition of neddylation by 3-day MLN4924 treatment regime does not impair autophagic flux in mouse hearts and brains.Am J Cardiovasc Dis. 2017 Dec 20;7(6):134-150. eCollection 2017. Am J Cardiovasc Dis. 2017. PMID: 29348974 Free PMC article.
-
Induction of Proteasome Subunit Low Molecular Weight Protein (LMP)-2 Is Required to Induce Active Remodeling in Adult Rat Ventricular Cardiomyocytes.Med Sci (Basel). 2020 May 1;8(2):21. doi: 10.3390/medsci8020021. Med Sci (Basel). 2020. PMID: 32370048 Free PMC article.
-
COP9 Signalosome Suppresses RIPK1-RIPK3-Mediated Cardiomyocyte Necroptosis in Mice.Circ Heart Fail. 2020 Aug;13(8):e006996. doi: 10.1161/CIRCHEARTFAILURE.120.006996. Epub 2020 Jun 24. Circ Heart Fail. 2020. PMID: 32578441 Free PMC article.
References
-
- Willis MS, Patterson C. Proteotoxicity and cardiac dysfunction--alzheimer's disease of the heart? N Engl J Med. 2013;368:455–464. - PubMed
-
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. - PubMed
-
- Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, Li F, Gerdes AM, Wawrousek EF, Wang X. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97:1018–1026. - PubMed
-
- Liu J, Tang M, Mestril R, Wang X. Aberrant protein aggregation is essential for a mutant desmin to impair the proteolytic function of the ubiquitin-proteasome system in cardiomyocytes. J Mol Cell Cardiol. 2006;40:451–454. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

