Host genetic variation impacts microbiome composition across human body sites
- PMID: 26374288
- PMCID: PMC4570153
- DOI: 10.1186/s13059-015-0759-1
Host genetic variation impacts microbiome composition across human body sites
Abstract
Background: The composition of bacteria in and on the human body varies widely across human individuals, and has been associated with multiple health conditions. While microbial communities are influenced by environmental factors, some degree of genetic influence of the host on the microbiome is also expected. This study is part of an expanding effort to comprehensively profile the interactions between human genetic variation and the composition of this microbial ecosystem on a genome- and microbiome-wide scale.
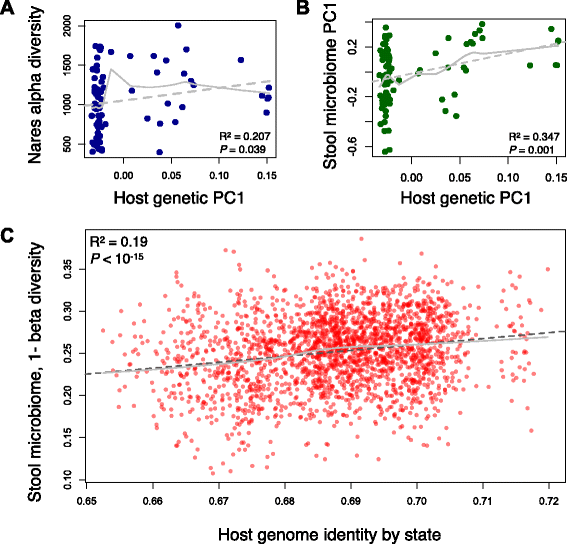
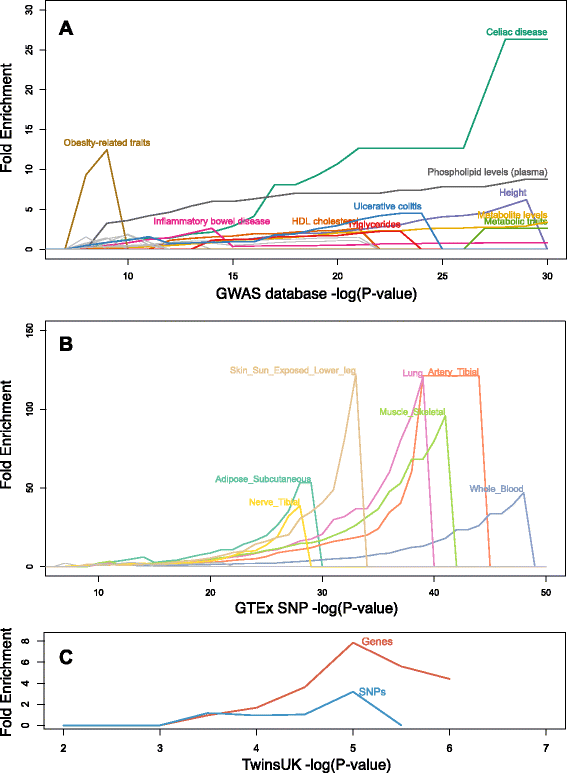
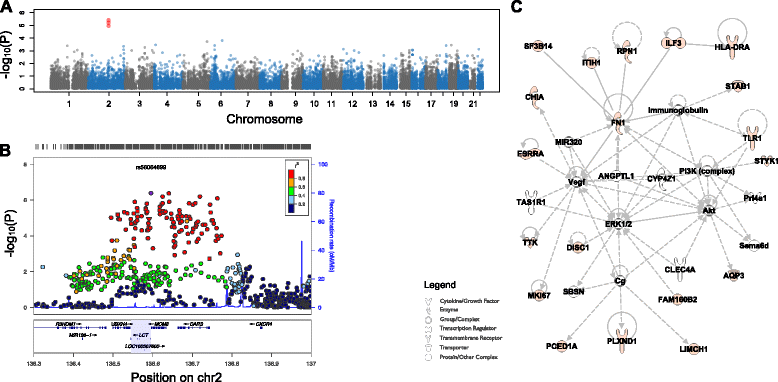
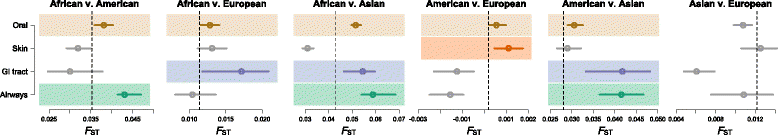
Results: Here, we jointly analyze the composition of the human microbiome and host genetic variation. By mining the shotgun metagenomic data from the Human Microbiome Project for host DNA reads, we gathered information on host genetic variation for 93 individuals for whom bacterial abundance data are also available. Using this dataset, we identify significant associations between host genetic variation and microbiome composition in 10 of the 15 body sites tested. These associations are driven by host genetic variation in immunity-related pathways, and are especially enriched in host genes that have been previously associated with microbiome-related complex diseases, such as inflammatory bowel disease and obesity-related disorders. Lastly, we show that host genomic regions associated with the microbiome have high levels of genetic differentiation among human populations, possibly indicating host genomic adaptation to environment-specific microbiomes.
Conclusions: Our results highlight the role of host genetic variation in shaping the composition of the human microbiome, and provide a starting point toward understanding the complex interaction between human genetics and the microbiome in the context of human evolution and disease.
Figures
Similar articles
-
The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome.mBio. 2015 Oct 20;6(5):e01578-15. doi: 10.1128/mBio.01578-15. mBio. 2015. PMID: 26489866 Free PMC article.
-
The East Asian gut microbiome is distinct from colocalized White subjects and connected to metabolic health.Elife. 2021 Oct 7;10:e70349. doi: 10.7554/eLife.70349. Elife. 2021. PMID: 34617511 Free PMC article.
-
Host genetic variation and its microbiome interactions within the Human Microbiome Project.Genome Med. 2018 Jan 29;10(1):6. doi: 10.1186/s13073-018-0515-8. Genome Med. 2018. PMID: 29378630 Free PMC article.
-
The role of animal hosts in shaping gut microbiome variation.Philos Trans R Soc Lond B Biol Sci. 2024 May 6;379(1901):20230071. doi: 10.1098/rstb.2023.0071. Epub 2024 Mar 18. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 38497257 Free PMC article. Review.
-
Elucidating the role of the host genome in shaping microbiome composition.Gut Microbes. 2016;7(2):178-84. doi: 10.1080/19490976.2016.1155022. Gut Microbes. 2016. PMID: 26939746 Free PMC article. Review.
Cited by
-
Gut microbiota, metabolites and host immunity.Nat Rev Immunol. 2016 May 27;16(6):341-52. doi: 10.1038/nri.2016.42. Nat Rev Immunol. 2016. PMID: 27231050 Free PMC article. Review.
-
Microbial and transcriptional differences elucidate atopic dermatitis heterogeneity across skin sites.Allergy. 2021 Apr;76(4):1173-1187. doi: 10.1111/all.14606. Epub 2020 Oct 14. Allergy. 2021. PMID: 33001460 Free PMC article.
-
Human Respiratory and Gut Microbiomes-Do They Really Contribute to Respiratory Health?Front Pediatr. 2020 Sep 3;8:528. doi: 10.3389/fped.2020.00528. eCollection 2020. Front Pediatr. 2020. PMID: 33014929 Free PMC article. Review.
-
A genome-wide association study reveals the relationship between human genetic variation and the nasal microbiome.Commun Biol. 2024 Jan 30;7(1):139. doi: 10.1038/s42003-024-05822-5. Commun Biol. 2024. PMID: 38291185 Free PMC article.
-
Acquisition and establishment of the oral microbiota.Periodontol 2000. 2021 Jun;86(1):123-141. doi: 10.1111/prd.12366. Epub 2021 Mar 10. Periodontol 2000. 2021. PMID: 33690935 Free PMC article. Review.
References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

