Decoding protein networks during virus entry by quantitative proteomics
- PMID: 26365680
- PMCID: PMC4914609
- DOI: 10.1016/j.virusres.2015.09.006
Decoding protein networks during virus entry by quantitative proteomics
Abstract
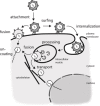
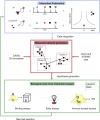
Virus entry into host cells relies on interactions between viral and host structures including lipids, carbohydrates and proteins. Particularly, protein-protein interactions between viral surface proteins and host proteins as well as secondary host protein-protein interactions play a pivotal role in coordinating virus binding and uptake. These interactions are dynamic and frequently involve multiprotein complexes. In the past decade mass spectrometry based proteomics methods have reached sensitivities and high throughput compatibilities of genomics methods and now allow the reliable quantitation of proteins in complex samples from limited material. As proteomics provides essential information on the biologically active entity namely the protein, including its posttranslational modifications and its interactions with other proteins, it is an indispensable method in the virologist's toolbox. Here we review protein interactions during virus entry and compare classical biochemical methods to study entry with novel technically advanced quantitative proteomics techniques. We highlight the value of quantitative proteomics in mapping functional virus entry networks, discuss the benefits and limitations and illustrate how the methodology will help resolve unsettled questions in virus entry research in the future.
Keywords: Protein interaction networks; Protein–protein interactions; Quantitative proteomics; Virus entry.
Copyright © 2015 The Authors. Published by Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Hunting Viral Receptors Using Haploid Cells.Annu Rev Virol. 2015 Nov;2(1):219-39. doi: 10.1146/annurev-virology-100114-055119. Epub 2015 Jul 2. Annu Rev Virol. 2015. PMID: 26958914 Free PMC article. Review.
-
Role of phosphatidylserine receptors in enveloped virus infection.J Virol. 2014 Apr;88(8):4275-90. doi: 10.1128/JVI.03287-13. Epub 2014 Jan 29. J Virol. 2014. PMID: 24478428 Free PMC article.
-
Viral proteomics: global evaluation of viruses and their interaction with the host.Expert Rev Proteomics. 2007 Dec;4(6):815-29. doi: 10.1586/14789450.4.6.815. Expert Rev Proteomics. 2007. PMID: 18067418 Review.
-
Beyond RGD: virus interactions with integrins.Arch Virol. 2015 Nov;160(11):2669-81. doi: 10.1007/s00705-015-2579-8. Epub 2015 Sep 1. Arch Virol. 2015. PMID: 26321473 Free PMC article. Review.
-
The C type lectins DC-SIGN and L-SIGN: receptors for viral glycoproteins.Methods Mol Biol. 2007;379:51-68. doi: 10.1007/978-1-59745-393-6_4. Methods Mol Biol. 2007. PMID: 17502670 Free PMC article. Review.
Cited by
-
Viroporins Manipulate Cellular Powerhouses and Modulate Innate Immunity.Viruses. 2024 Feb 23;16(3):345. doi: 10.3390/v16030345. Viruses. 2024. PMID: 38543711 Free PMC article. Review.
-
Dynamic Dystroglycan Complexes Mediate Cell Entry of Lassa Virus.mBio. 2019 Mar 26;10(2):e02869-18. doi: 10.1128/mBio.02869-18. mBio. 2019. PMID: 30914516 Free PMC article.
-
Genome assembly of Stephania longa provides insight into cepharanthine biosynthesis.Front Plant Sci. 2024 Sep 5;15:1414636. doi: 10.3389/fpls.2024.1414636. eCollection 2024. Front Plant Sci. 2024. PMID: 39301160 Free PMC article.
-
Hepatitis C Virus Entry: Protein Interactions and Fusion Determinants Governing Productive Hepatocyte Invasion.Cold Spring Harb Perspect Med. 2020 Feb 3;10(2):a036830. doi: 10.1101/cshperspect.a036830. Cold Spring Harb Perspect Med. 2020. PMID: 31427285 Free PMC article. Review.
-
Dual Analysis of Virus-Host Interactions: The Case of Ostreid herpesvirus 1 and the Cupped Oyster Crassostrea gigas.Evol Bioinform Online. 2019 Feb 22;15:1176934319831305. doi: 10.1177/1176934319831305. eCollection 2019. Evol Bioinform Online. 2019. PMID: 30828244 Free PMC article. Review.
References
-
- Aloy P., Russell R.B. Ten thousand interactions for the molecular biologist. Nat. Biotechnol. 2004;22:1317–1321. - PubMed
-
- Andersen J.S., Matic I., Vertegaal A.C. Identification of SUMO target proteins by quantitative proteomics. Methods Mol. Biol. 2009;497:19–31. - PubMed
-
- Berard A.R., Coombs K.M., Severini A. Quantification of the host response proteome after herpes simplex virus type 1 infection. J. Proteome Res. 2015;14:2121–2142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

