Genome-wide target analysis of NEUROD2 provides new insights into regulation of cortical projection neuron migration and differentiation
- PMID: 26341353
- PMCID: PMC4560887
- DOI: 10.1186/s12864-015-1882-9
Genome-wide target analysis of NEUROD2 provides new insights into regulation of cortical projection neuron migration and differentiation
Abstract
Background: Cellular differentiation programs are controlled, to a large extent, by the combinatorial functioning of specific transcription factors. Cortical projection neurons constitute the major excitatory neuron population within the cortex and mediate long distance communication between the cortex and other brain regions. Our understanding of effector transcription factors and their downstream transcriptional programs that direct the differentiation process of cortical projection neurons is far from complete.
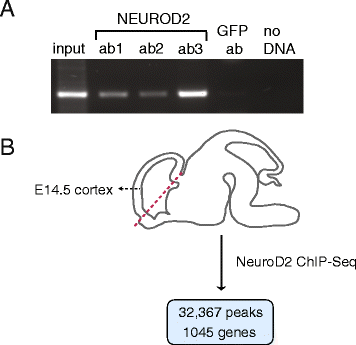
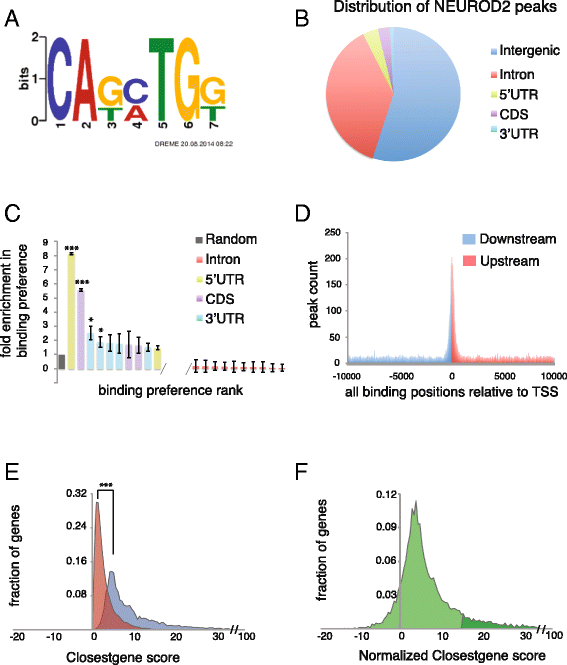
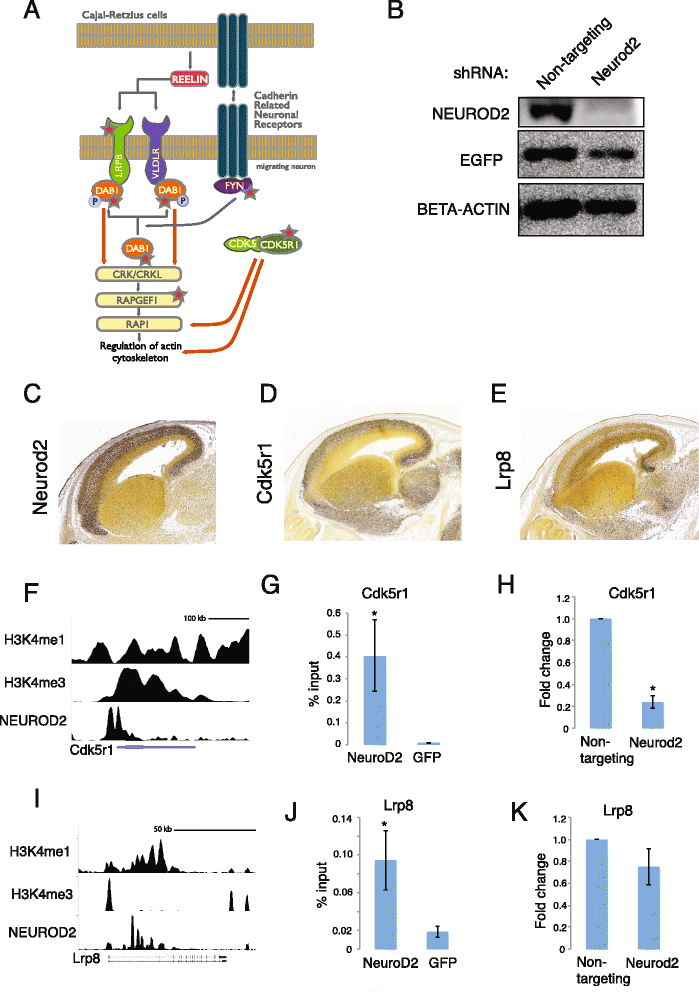
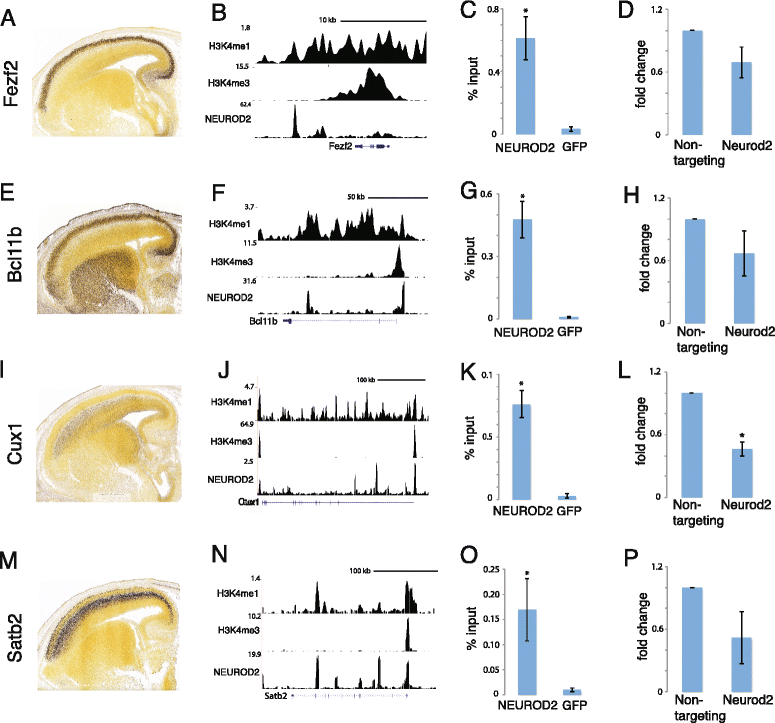
Results: In this study, we carried out a ChIP-Seq (chromatin-immunoprecipitation and sequencing) analysis of NEUROD2, an effector transcription factor expressed in lineages of cortical projection neurons during the peak of cortical excitatory neurogenesis. Our results suggest that during cortical development NEUROD2 targets key genes that are required for Reelin signaling, a major pathway that regulates the migration of neurons from germinal zones to their final layers of residence within the cortex. We also find that NEUROD2 binds to a large set of genes with functions in layer-specific differentiation and in axonal pathfinding of cortical projection neurons.
Conclusions: Our analysis of in vivo NEUROD2 target genes offers mechanistic insight into signaling pathways that regulate neuronal migration and axon guidance and identifies genes that are likely to be required for proper cortical development.
Figures

Similar articles
-
Terminal neuron localization to the upper cortical plate is controlled by the transcription factor NEUROD2.Sci Rep. 2019 Dec 23;9(1):19697. doi: 10.1038/s41598-019-56171-x. Sci Rep. 2019. PMID: 31873146 Free PMC article.
-
The E2A splice variant E47 regulates the differentiation of projection neurons via p57(KIP2) during cortical development.Development. 2017 Nov 1;144(21):3917-3931. doi: 10.1242/dev.145698. Epub 2017 Sep 22. Development. 2017. PMID: 28939666
-
The transcription factor NeuroD2 coordinates synaptic innervation and cell intrinsic properties to control excitability of cortical pyramidal neurons.J Physiol. 2016 Jul 1;594(13):3729-44. doi: 10.1113/JP271953. J Physiol. 2016. PMID: 27146976 Free PMC article.
-
From radial glia to pyramidal-projection neuron: transcription factor cascades in cerebral cortex development.Mol Neurobiol. 2006 Feb;33(1):33-50. doi: 10.1385/MN:33:1:033. Mol Neurobiol. 2006. PMID: 16388109 Review.
-
Molecular pathways controlling the sequential steps of cortical projection neuron migration.Adv Exp Med Biol. 2014;800:1-24. doi: 10.1007/978-94-007-7687-6_1. Adv Exp Med Biol. 2014. PMID: 24243097 Review.
Cited by
-
Maternal hyperglycemia disturbs neocortical neurogenesis via epigenetic regulation in C57BL/6J mice.Cell Death Dis. 2019 Mar 1;10(3):211. doi: 10.1038/s41419-019-1438-z. Cell Death Dis. 2019. PMID: 30824686 Free PMC article.
-
miR-153 attenuates the inflammatory response and oxidative stress induced by spinal cord injury by targeting of NEUROD2.Am J Transl Res. 2021 Jul 15;13(7):7968-7975. eCollection 2021. Am J Transl Res. 2021. PMID: 34377277 Free PMC article.
-
Multiscale 3D Genome Rewiring during Mouse Neural Development.Cell. 2017 Oct 19;171(3):557-572.e24. doi: 10.1016/j.cell.2017.09.043. Cell. 2017. PMID: 29053968 Free PMC article.
-
Deciphering the distinct transcriptomic and gene regulatory map in adult macaque basal ganglia cells.Gigascience. 2022 Dec 28;12:giad095. doi: 10.1093/gigascience/giad095. Epub 2023 Dec 13. Gigascience. 2022. PMID: 38091510 Free PMC article.
-
Arc Regulates Transcription of Genes for Plasticity, Excitability and Alzheimer's Disease.Biomedicines. 2022 Aug 11;10(8):1946. doi: 10.3390/biomedicines10081946. Biomedicines. 2022. PMID: 36009494 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

