Activity dependent CAM cleavage and neurotransmission
- PMID: 26321910
- PMCID: PMC4531370
- DOI: 10.3389/fncel.2015.00305
Activity dependent CAM cleavage and neurotransmission
Abstract
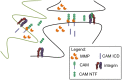
Spatially localized proteolysis represents an elegant means by which neuronal activity dependent changes in synaptic structure, and thus experience dependent learning and memory, can be achieved. In vitro and in vivo studies suggest that matrix metalloproteinase and adamalysin activity is concentrated at the cell surface, and emerging evidence suggests that increased peri-synaptic expression, release and/or activation of these proteinases occurs with enhanced excitatory neurotransmission. Synaptically expressed cell adhesion molecules (CAMs) could therefore represent important targets for neuronal activity-dependent proteolysis. Several CAM subtypes are expressed at the synapse, and their cleavage can influence the efficacy of synaptic transmission through a variety of non-mutually exclusive mechanisms. In the following review, we discuss mechanisms that regulate neuronal activity-dependent synaptic CAM shedding, including those that may be calcium dependent. We also highlight CAM targets of activity-dependent proteolysis including neuroligin and intercellular adhesion molecule-5 (ICAM-5). We include discussion focused on potential consequences of synaptic CAM shedding, with an emphasis on interactions between soluble CAM cleavage products and specific pre- and post-synaptic receptors.
Keywords: CAM; MMP; adhesion; dendritic spine; glutamate; metalloproteases.
Figures
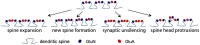
Similar articles
-
MMPs and soluble ICAM-5 increase neuronal excitability within in vitro networks of hippocampal neurons.PLoS One. 2012;7(8):e42631. doi: 10.1371/journal.pone.0042631. Epub 2012 Aug 13. PLoS One. 2012. PMID: 22912716 Free PMC article.
-
Methamphetamine-associated cleavage of the synaptic adhesion molecule intercellular adhesion molecule-5.J Neurochem. 2011 Aug;118(4):521-32. doi: 10.1111/j.1471-4159.2010.07153.x. Epub 2011 Jan 19. J Neurochem. 2011. PMID: 21166806 Free PMC article.
-
Soluble Ectodomain of Neuroligin 1 Decreases Synaptic Activity by Activating Metabotropic Glutamate Receptor 2.Front Mol Neurosci. 2017 May 3;10:116. doi: 10.3389/fnmol.2017.00116. eCollection 2017. Front Mol Neurosci. 2017. PMID: 28515678 Free PMC article.
-
Cell adhesion and homeostatic synaptic plasticity.Neuropharmacology. 2014 Mar;78:23-30. doi: 10.1016/j.neuropharm.2013.03.015. Epub 2013 Mar 28. Neuropharmacology. 2014. PMID: 23542441 Review.
-
Calmodulin-dependent protein kinase II. Multifunctional roles in neuronal differentiation and synaptic plasticity.Mol Neurobiol. 1991;5(2-4):153-77. doi: 10.1007/BF02935544. Mol Neurobiol. 1991. PMID: 1668384 Review.
Cited by
-
GSK-3β and MMP-9 Cooperate in the Control of Dendritic Spine Morphology.Mol Neurobiol. 2017 Jan;54(1):200-211. doi: 10.1007/s12035-015-9625-0. Epub 2016 Jan 6. Mol Neurobiol. 2017. PMID: 26738851 Free PMC article.
-
Proteolytic cleavage is required for functional neuroligin 2 maturation and trafficking in Drosophila.J Mol Cell Biol. 2017 Jun 1;9(3):231-242. doi: 10.1093/jmcb/mjx015. J Mol Cell Biol. 2017. PMID: 28498949 Free PMC article.
-
Shed CNTNAP2 ectodomain is detectable in CSF and regulates Ca2+ homeostasis and network synchrony via PMCA2/ATP2B2.Neuron. 2022 Feb 16;110(4):627-643.e9. doi: 10.1016/j.neuron.2021.11.025. Epub 2021 Dec 17. Neuron. 2022. PMID: 34921780 Free PMC article.
-
Diagnostic and Therapeutic Approaches for Glioblastoma and Neuroblastoma Cancers Using Chlorotoxin Nanoparticles.Cancers (Basel). 2023 Jun 28;15(13):3388. doi: 10.3390/cancers15133388. Cancers (Basel). 2023. PMID: 37444498 Free PMC article. Review.
-
Matrix disequilibrium in Alzheimer's disease and conditions that increase Alzheimer's disease risk.Front Neurosci. 2023 May 26;17:1188065. doi: 10.3389/fnins.2023.1188065. eCollection 2023. Front Neurosci. 2023. PMID: 37304012 Free PMC article. Review.
References
-
- Anderson G. R., Galfin T., Xu W., Aoto J., Malenka R. C., Sudhof T. C. (2012). Candidate autism gene screen identifies critical role for cell-adhesion molecule CASPR2 in dendritic arborization and spine development. Proc. Natl. Acad. Sci. U.S.A. 109 18120–18125. 10.1073/pnas.1216398109 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

