Analysis of Small Ubiquitin-Like Modifier (SUMO) Targets Reflects the Essential Nature of Protein SUMOylation and Provides Insight to Elucidate the Role of SUMO in Plant Development
- PMID: 26320229
- PMCID: PMC4587472
- DOI: 10.1104/pp.15.01014
Analysis of Small Ubiquitin-Like Modifier (SUMO) Targets Reflects the Essential Nature of Protein SUMOylation and Provides Insight to Elucidate the Role of SUMO in Plant Development
Abstract
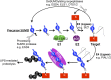
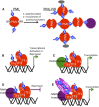
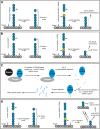
Posttranslational modification of proteins by small ubiquitin-like modifier (SUMO) has received much attention, reflected by a flood of recent studies implicating SUMO in a wide range of cellular and molecular activities, many of which are conserved throughout eukaryotes. Whereas most of these studies were performed in vitro or in single cells, plants provide an excellent system to study the role of SUMO at the developmental level. Consistent with its essential roles during plant development, mutations of the basic SUMOylation machinery in Arabidopsis (Arabidopsis thaliana) cause embryo stage arrest or major developmental defects due to perturbation of the dynamics of target SUMOylation. Efforts to identify SUMO protein targets in Arabidopsis have been modest; however, recent success in identifying thousands of human SUMO targets using unique experimental designs can potentially help identify plant SUMO targets more efficiently. Here, known Arabidopsis SUMO targets are reevaluated, and potential approaches to dissect the roles of SUMO in plant development are discussed.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures

Similar articles
-
New insights into the role of the small ubiquitin-like modifier (SUMO) in plants.Int Rev Cell Mol Biol. 2013;300:161-209. doi: 10.1016/B978-0-12-405210-9.00005-9. Int Rev Cell Mol Biol. 2013. PMID: 23273862 Review.
-
mRNA export and sumoylation-Lessons from plants.Biochim Biophys Acta. 2012 Jun;1819(6):531-7. doi: 10.1016/j.bbagrm.2012.01.006. Epub 2012 Jan 28. Biochim Biophys Acta. 2012. PMID: 22306659 Review.
-
Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry.Plant Cell Physiol. 2009 Jun;50(6):1049-61. doi: 10.1093/pcp/pcp056. Epub 2009 Apr 17. Plant Cell Physiol. 2009. PMID: 19376783
-
SIZ1-Dependent Post-Translational Modification by SUMO Modulates Sugar Signaling and Metabolism in Arabidopsis thaliana.Plant Cell Physiol. 2015 Dec;56(12):2297-311. doi: 10.1093/pcp/pcv149. Epub 2015 Oct 14. Plant Cell Physiol. 2015. PMID: 26468507
-
The Ubiquitin-Like SUMO System and Heart Function: From Development to Disease.Circ Res. 2016 Jan 8;118(1):132-44. doi: 10.1161/CIRCRESAHA.115.307730. Circ Res. 2016. PMID: 26837744 Review.
Cited by
-
Genome-wide profiling of histone (H3) lysine 4 (K4) tri-methylation (me3) under drought, heat, and combined stresses in switchgrass.BMC Genomics. 2024 Feb 29;25(1):223. doi: 10.1186/s12864-024-10068-w. BMC Genomics. 2024. PMID: 38424499 Free PMC article.
-
Deciphering the intricate hierarchical gene regulatory network: unraveling multi-level regulation and modifications driving secondary cell wall formation.Hortic Res. 2023 Dec 19;11(2):uhad281. doi: 10.1093/hr/uhad281. eCollection 2024 Feb. Hortic Res. 2023. PMID: 38344650 Free PMC article.
-
The converging path of protein SUMOylation in phytohormone signalling: highlights and new frontiers.Plant Cell Rep. 2021 Nov;40(11):2047-2061. doi: 10.1007/s00299-021-02732-2. Epub 2021 Jun 15. Plant Cell Rep. 2021. PMID: 34129078 Review.
-
Protein sumoylation and phosphorylation intersect in Arabidopsis signaling.Plant J. 2017 Aug;91(3):505-517. doi: 10.1111/tpj.13575. Epub 2017 Jun 4. Plant J. 2017. PMID: 28419593 Free PMC article.
-
Phototropin Interactions with SUMO Proteins.Plant Cell Physiol. 2021 Sep 24;62(4):693-707. doi: 10.1093/pcp/pcab027. Plant Cell Physiol. 2021. PMID: 33594440 Free PMC article.
References
-
- Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M (2005) Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 435: 979–982 - PubMed
-
- Becker J, Barysch SV, Karaca S, Dittner C, Hsiao HH, Berriel Diaz M, Herzig S, Urlaub H, Melchior F (2013) Detecting endogenous SUMO targets in mammalian cells and tissues. Nat Struct Mol Biol 20: 525–531 - PubMed
-
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

