Replication-Competent Influenza Virus and Respiratory Syncytial Virus Luciferase Reporter Strains Engineered for Co-Infections Identify Antiviral Compounds in Combination Screens
- PMID: 26307636
- PMCID: PMC4719150
- DOI: 10.1021/acs.biochem.5b00623
Replication-Competent Influenza Virus and Respiratory Syncytial Virus Luciferase Reporter Strains Engineered for Co-Infections Identify Antiviral Compounds in Combination Screens
Abstract
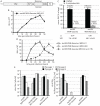
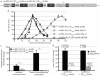
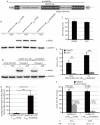
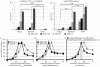
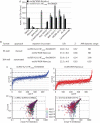
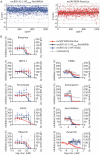
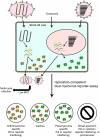
Myxoviruses such as influenza A virus (IAV) and respiratory syncytial virus (RSV) are major human pathogens, mandating the development of novel therapeutics. To establish a high-throughput screening protocol for the simultaneous identification of pathogen- and host-targeted hit candidates against either pathogen or both, we have attempted co-infection of cells with IAV and RSV. However, viral replication kinetics were incompatible, RSV signal window was low, and an IAV-driven minireplicon reporter assay used in initial screens narrowed the host cell range and restricted the assay to single-cycle infections. To overcome these limitations, we developed an RSV strain carrying firefly luciferase fused to an innovative universal small-molecule assisted shut-off domain, which boosted assay signal window, and a hyperactive fusion protein that synchronized IAV and RSV reporter expression kinetics and suppressed the identification of RSV entry inhibitors sensitive to a recently reported RSV pan-resistance mechanism. Combined with a replication-competent recombinant IAV strain harboring nanoluciferase, the assay performed well on a human respiratory cell line and supports multicycle infections. Miniaturized to 384-well format, the protocol was validated through screening of a set of the National Institutes of Health Clinical Collection (NCC) in quadruplicate. These test screens demonstrated favorable assay parameters and reproducibility. Application to a LOPAC library of bioactive compounds in a proof-of-concept campaign detected licensed antimyxovirus therapeutics, ribavirin and the neuraminidase inhibitor zanamivir, and identified two unexpected RSV-specific hit candidates, Fenretinide and the opioid receptor antagonist BNTX-7. Hits were evaluated in direct and orthogonal dose-response counterscreens using a standard recRSV reporter strain expressing Renilla luciferase.
Figures
Similar articles
-
Replication-Competent Influenza A Viruses Expressing Reporter Genes.Viruses. 2016 Jun 23;8(7):179. doi: 10.3390/v8070179. Viruses. 2016. PMID: 27347991 Free PMC article. Review.
-
Identification and Characterization of Influenza Virus Entry Inhibitors through Dual Myxovirus High-Throughput Screening.J Virol. 2016 Jul 27;90(16):7368-7387. doi: 10.1128/JVI.00898-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27252534 Free PMC article.
-
Dual myxovirus screen identifies a small-molecule agonist of the host antiviral response.J Virol. 2013 Oct;87(20):11076-87. doi: 10.1128/JVI.01425-13. Epub 2013 Aug 7. J Virol. 2013. PMID: 23926334 Free PMC article.
-
A Novel Fluorescent and Bioluminescent Bireporter Influenza A Virus To Evaluate Viral Infections.J Virol. 2019 May 1;93(10):e00032-19. doi: 10.1128/JVI.00032-19. Print 2019 May 15. J Virol. 2019. PMID: 30867298 Free PMC article.
-
Drug candidates and model systems in respiratory syncytial virus antiviral drug discovery.Biochem Pharmacol. 2017 Mar 1;127:1-12. doi: 10.1016/j.bcp.2016.09.014. Epub 2016 Sep 19. Biochem Pharmacol. 2017. PMID: 27659812 Review.
Cited by
-
Recombinant Influenza A Viruses Expressing Reporter Genes from the Viral NS Segment.Int J Mol Sci. 2024 Oct 1;25(19):10584. doi: 10.3390/ijms251910584. Int J Mol Sci. 2024. PMID: 39408912 Free PMC article. Review.
-
Generation of a pdmH1N1 2018 Influenza A Reporter Virus Carrying a mCherry Fluorescent Protein in the PA Segment.Front Cell Infect Microbiol. 2022 Jan 20;11:827790. doi: 10.3389/fcimb.2021.827790. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35127568 Free PMC article.
-
Segmented Filamentous Bacteria Prevent and Cure Rotavirus Infection.Cell. 2019 Oct 17;179(3):644-658.e13. doi: 10.1016/j.cell.2019.09.028. Epub 2019 Oct 10. Cell. 2019. PMID: 31607511 Free PMC article.
-
Orally efficacious lead of the AVG inhibitor series targeting a dynamic interface in the respiratory syncytial virus polymerase.Sci Adv. 2022 Jun 24;8(25):eabo2236. doi: 10.1126/sciadv.abo2236. Epub 2022 Jun 24. Sci Adv. 2022. PMID: 35749502 Free PMC article.
-
Replication-Competent Influenza A Viruses Expressing Reporter Genes.Viruses. 2016 Jun 23;8(7):179. doi: 10.3390/v8070179. Viruses. 2016. PMID: 27347991 Free PMC article. Review.
References
-
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–186. - PubMed
-
- Hall CB. Respiratory syncytial virus and parainfluenza virus. The New England journal of medicine. 2001;344:1917–1928. - PubMed
-
- Mahadevia PJ, Masaquel AS, Polak MJ, Weiner LB. Cost utility of palivizumab prophylaxis among pre-term infants in the United States: a national policy perspective. J Med Econ. 2012;15:987–996. - PubMed
-
- Collins PL, Crowe JE., Jr. Respiratory Syncytial Virus and Metapneumoviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5 Lippincott, Williams, & Wilkins; Philadelphia: 2007. pp. 1601–1645.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

