Integrated Transcriptomics Establish Macrophage Polarization Signatures and have Potential Applications for Clinical Health and Disease
- PMID: 26302899
- PMCID: PMC4548187
- DOI: 10.1038/srep13351
Integrated Transcriptomics Establish Macrophage Polarization Signatures and have Potential Applications for Clinical Health and Disease
Abstract
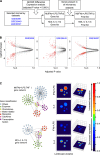
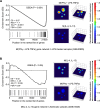
Growing evidence defines macrophages (Mφ) as plastic cells with wide-ranging states of activation and expression of different markers that are time and location dependent. Distinct from the simple M1/M2 dichotomy initially proposed, extensive diversity of macrophage phenotypes have been extensively demonstrated as characteristic features of monocyte-macrophage differentiation, highlighting the difficulty of defining complex profiles by a limited number of genes. Since the description of macrophage activation is currently contentious and confusing, the generation of a simple and reliable framework to categorize major Mφ phenotypes in the context of complex clinical conditions would be extremely relevant to unravel different roles played by these cells in pathophysiological scenarios. In the current study, we integrated transcriptome data using bioinformatics tools to generate two macrophage molecular signatures. We validated our signatures in in vitro experiments and in clinical samples. More importantly, we were able to attribute prognostic and predictive values to components of our signatures. Our study provides a framework to guide the interrogation of macrophage phenotypes in the context of health and disease. The approach described here could be used to propose new biomarkers for diagnosis in diverse clinical settings including dengue infections, asthma and sepsis resolution.
Figures


Similar articles
-
Transcriptional and functional diversity of human macrophage repolarization.J Allergy Clin Immunol. 2019 Apr;143(4):1536-1548. doi: 10.1016/j.jaci.2018.10.046. Epub 2018 Nov 14. J Allergy Clin Immunol. 2019. PMID: 30445062 Free PMC article.
-
Trajectory analysis quantifies transcriptional plasticity during macrophage polarization.Sci Rep. 2020 Jul 23;10(1):12273. doi: 10.1038/s41598-020-68766-w. Sci Rep. 2020. PMID: 32703960 Free PMC article.
-
Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression.J Immunol. 2006 Nov 15;177(10):7303-11. doi: 10.4049/jimmunol.177.10.7303. J Immunol. 2006. PMID: 17082649
-
Classical Dichotomy of Macrophages and Alternative Activation Models Proposed with Technological Progress.Biomed Res Int. 2021 Oct 21;2021:9910596. doi: 10.1155/2021/9910596. eCollection 2021. Biomed Res Int. 2021. PMID: 34722776 Free PMC article. Review.
-
Implications of macrophage polarization in autoimmunity.Immunology. 2018 Jun;154(2):186-195. doi: 10.1111/imm.12910. Epub 2018 Mar 8. Immunology. 2018. PMID: 29455468 Free PMC article. Review.
Cited by
-
Eschar dissolution and the immunoregulator effect of keratinase on burn wounds.Sci Rep. 2023 Aug 14;13(1):13238. doi: 10.1038/s41598-023-39765-4. Sci Rep. 2023. PMID: 37580372 Free PMC article.
-
Dectin-1 stimulation promotes a distinct inflammatory signature in the setting of HIV-infection and aging.Aging (Albany NY). 2023 Aug 21;15(16):7866-7908. doi: 10.18632/aging.204927. Epub 2023 Aug 21. Aging (Albany NY). 2023. PMID: 37606991 Free PMC article.
-
Descriptive vs mechanistic scientific approach to study wound healing and its inhibition: Is there a value of translational research involving human subjects?Exp Dermatol. 2018 May;27(5):551-562. doi: 10.1111/exd.13663. Exp Dermatol. 2018. PMID: 29660181 Free PMC article. Review.
-
Attenuated inflammatory response of monocyte-derived macrophage from patients with BD: a preliminary report.Int J Bipolar Disord. 2019 Jun 1;7(1):13. doi: 10.1186/s40345-019-0148-x. Int J Bipolar Disord. 2019. PMID: 31152269 Free PMC article.
-
Immune cell profiling in the age of immune checkpoint inhibitors: implications for biomarker discovery and understanding of resistance mechanisms.Mamm Genome. 2018 Dec;29(11-12):866-878. doi: 10.1007/s00335-018-9757-4. Epub 2018 Jul 2. Mamm Genome. 2018. PMID: 29968076 Free PMC article. Review.
References
-
- Biswas S. K., Chittezhath M., Shalova I. N. & Lim J.-Y. Macrophage polarization and plasticity in health and disease. Immunol Res 53, 11–24 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

