Antisense-mediated affinity purification of dengue virus ribonucleoprotein complexes from infected cells
- PMID: 26276314
- PMCID: PMC4684824
- DOI: 10.1016/j.ymeth.2015.08.008
Antisense-mediated affinity purification of dengue virus ribonucleoprotein complexes from infected cells
Abstract
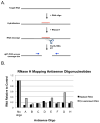
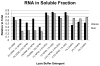
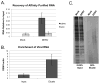
The identification of RNA-binding proteins that physically associate with viral RNA molecules during infection can provide insight into the molecular mechanisms of RNA virus replication. Until recently, such RNA-protein interactions have been identified predominantly with the use of in vitro assays that may not accurately reflect associations that occur in the context of a living cell. Here we describe a method for the specific affinity purification of dengue virus RNA and associated proteins using in vivo cross-linking followed by antisense-mediated affinity purification. RNA-binding proteins that specifically co-purify with viral RNA using this method can be identified en masse by mass spectrometry. This strategy can potentially be adapted to the purification of any viral RNA species of interest.
Keywords: Dengue virus; RNA affinity chromatography; Ribonucleoprotein complex.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
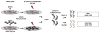
Similar articles
-
Identification of Proteins Bound to Dengue Viral RNA In Vivo Reveals New Host Proteins Important for Virus Replication.mBio. 2016 Jan 5;7(1):e01865-15. doi: 10.1128/mBio.01865-15. mBio. 2016. PMID: 26733069 Free PMC article.
-
Isolation of Endogenously Assembled RNA-Protein Complexes Using Affinity Purification Based on Streptavidin Aptamer S1.Int J Mol Sci. 2015 Sep 16;16(9):22456-72. doi: 10.3390/ijms160922456. Int J Mol Sci. 2015. PMID: 26389898 Free PMC article.
-
Isolation of Cognate Cellular and Viral Ribonucleoprotein Complexes of HIV-1 RNA Applicable to Proteomic Discovery and Molecular Investigations.Methods Mol Biol. 2016;1354:133-46. doi: 10.1007/978-1-4939-3046-3_9. Methods Mol Biol. 2016. PMID: 26714709
-
Isolation of viral ribonucleoprotein complexes from infected cells by tandem affinity purification.Proteomics. 2005 Nov;5(17):4483-7. doi: 10.1002/pmic.200402095. Proteomics. 2005. PMID: 16206331
-
Isolation and characterization of RNA-binding proteins from Drosophila melanogaster.Methods Cell Biol. 1994;44:191-205. doi: 10.1016/s0091-679x(08)60914-0. Methods Cell Biol. 1994. PMID: 7707952 Review. No abstract available.
Cited by
-
Revealing the Arabidopsis AtGRP7 mRNA binding proteome by specific enhanced RNA interactome capture.BMC Plant Biol. 2024 Jun 14;24(1):552. doi: 10.1186/s12870-024-05249-4. BMC Plant Biol. 2024. PMID: 38877390 Free PMC article.
-
Identification of Proteins Bound to Dengue Viral RNA In Vivo Reveals New Host Proteins Important for Virus Replication.mBio. 2016 Jan 5;7(1):e01865-15. doi: 10.1128/mBio.01865-15. mBio. 2016. PMID: 26733069 Free PMC article.
-
A widely applicable and cost-effective method for specific RNA-protein complex isolation.Sci Rep. 2023 Apr 27;13(1):6898. doi: 10.1038/s41598-023-34157-0. Sci Rep. 2023. PMID: 37106019 Free PMC article.
-
RNA-Centric Approaches to Profile the RNA-Protein Interaction Landscape on Selected RNAs.Noncoding RNA. 2021 Feb 15;7(1):11. doi: 10.3390/ncrna7010011. Noncoding RNA. 2021. PMID: 33671874 Free PMC article. Review.
-
Involvement of ribosomal protein L17 and Y-box binding protein 1 in the assembly of hepatitis C virus potentially via their interaction with the 3' untranslated region of the viral genome.J Virol. 2024 Jul 23;98(7):e0052224. doi: 10.1128/jvi.00522-24. Epub 2024 Jun 20. J Virol. 2024. PMID: 38899899 Free PMC article.
References
-
- Deo S, et al. Characterization of the termini of the West Nile virus genome and their interactions with the small isoform of the 2′ 5′-oligoadenylate synthetase family. J Struct Biol. 2015;190(2):236–49. - PubMed
-
- Paranjape SM, Harris E. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J Biol Chem. 2007;282(42):30497–508. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

