A Molecular Staple: D-Loops in the I Domain of Bacteriophage P22 Coat Protein Make Important Intercapsomer Contacts Required for Procapsid Assembly
- PMID: 26269173
- PMCID: PMC4580156
- DOI: 10.1128/JVI.01629-15
A Molecular Staple: D-Loops in the I Domain of Bacteriophage P22 Coat Protein Make Important Intercapsomer Contacts Required for Procapsid Assembly
Abstract
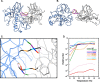
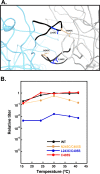
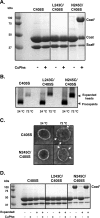
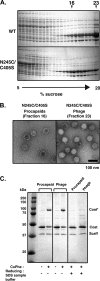
Bacteriophage P22, a double-stranded DNA (dsDNA) virus, has a nonconserved 124-amino-acid accessory domain inserted into its coat protein, which has the canonical HK97 protein fold. This I domain is involved in virus capsid size determination and stability, as well as protein folding. The nuclear magnetic resonance (NMR) solution structure of the I domain revealed the presence of a D-loop, which was hypothesized to make important intersubunit contacts between coat proteins in adjacent capsomers. Here we show that amino acid substitutions of residues near the tip of the D-loop result in aberrant assembly products, including tubes and broken particles, highlighting the significance of the D-loops in proper procapsid assembly. Using disulfide cross-linking, we showed that the tips of the D-loops are positioned directly across from each other both in the procapsid and the mature virion, suggesting their importance in both states. Our results indicate that D-loop interactions act as "molecular staples" at the icosahedral 2-fold symmetry axis and significantly contribute to stabilizing the P22 capsid for DNA packaging.
Importance: Many dsDNA viruses have morphogenic pathways utilizing an intermediate capsid, known as a procapsid. These procapsids are assembled from a coat protein having the HK97 fold in a reaction driven by scaffolding proteins or delta domains. Maturation of the capsid occurs during DNA packaging. Bacteriophage HK97 uniquely stabilizes its capsid during maturation by intercapsomer cross-linking, but most virus capsids are stabilized by alternate means. Here we show that the I domain that is inserted into the coat protein of bacteriophage P22 is important in the process of proper procapsid assembly. Specifically, the I domain allows for stabilizing interactions across the capsid 2-fold axis of symmetry via a D-loop. When amino acid residues at the tip of the D-loop are mutated, aberrant assembly products, including tubes, are formed instead of procapsids, consequently phage production is affected, indicating the importance of stabilizing interactions during the assembly and maturation reactions.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Of capsid structure and stability: The partnership between charged residues of E-loop and P-domain of the bacteriophage P22 coat protein.Virology. 2019 Aug;534:45-53. doi: 10.1016/j.virol.2019.05.021. Epub 2019 Jun 2. Virology. 2019. PMID: 31176063 Free PMC article.
-
Domain study of bacteriophage p22 coat protein and characterization of the capsid lattice transformation by hydrogen/deuterium exchange.J Mol Biol. 2005 Apr 15;347(5):935-48. doi: 10.1016/j.jmb.2005.02.021. J Mol Biol. 2005. PMID: 15784254
-
Highly specific salt bridges govern bacteriophage P22 icosahedral capsid assembly: identification of the site in coat protein responsible for interaction with scaffolding protein.J Virol. 2014 May;88(10):5287-97. doi: 10.1128/JVI.00036-14. Epub 2014 Mar 5. J Virol. 2014. PMID: 24600011 Free PMC article.
-
'Let the phage do the work': using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants.Virology. 2010 Jun 5;401(2):119-30. doi: 10.1016/j.virol.2010.02.017. Epub 2010 Mar 16. Virology. 2010. PMID: 20236676 Free PMC article. Review.
-
Nature's favorite building block: Deciphering folding and capsid assembly of proteins with the HK97-fold.Virology. 2015 May;479-480:487-97. doi: 10.1016/j.virol.2015.02.055. Epub 2015 Apr 8. Virology. 2015. PMID: 25864106 Free PMC article. Review.
Cited by
-
NMR assignments for the insertion domain of bacteriophage Sf6 coat protein.Biomol NMR Assign. 2017 Apr;11(1):35-38. doi: 10.1007/s12104-016-9716-5. Epub 2016 Oct 31. Biomol NMR Assign. 2017. PMID: 27798771 Free PMC article.
-
Portal protein functions akin to a DNA-sensor that couples genome-packaging to icosahedral capsid maturation.Nat Commun. 2017 Jan 30;8:14310. doi: 10.1038/ncomms14310. Nat Commun. 2017. PMID: 28134243 Free PMC article.
-
The phage L capsid decoration protein has a novel OB-fold and an unusual capsid binding strategy.Elife. 2019 Apr 4;8:e45345. doi: 10.7554/eLife.45345. Elife. 2019. PMID: 30945633 Free PMC article.
-
Coat Protein Mutations That Alter the Flux of Morphogenetic Intermediates through the ϕX174 Early Assembly Pathway.J Virol. 2017 Nov 30;91(24):e01384-17. doi: 10.1128/JVI.01384-17. Print 2017 Dec 15. J Virol. 2017. PMID: 28978706 Free PMC article.
-
Templated trimerization of the phage L decoration protein on capsids.bioRxiv [Preprint]. 2024 Sep 8:2024.09.08.611893. doi: 10.1101/2024.09.08.611893. bioRxiv. 2024. PMID: 39282432 Free PMC article. Preprint.
References
-
- Casjens S, King J. 1974. P22 morphogenesis. I. Catalytic scaffolding protein in capsid assembly. J Supramol Struct 2:202–224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

