Antagonist Xist and Tsix co-transcription during mouse oogenesis and maternal Xist expression during pre-implantation development calls into question the nature of the maternal imprint on the X chromosome
- PMID: 26267271
- PMCID: PMC4844198
- DOI: 10.1080/15592294.2015.1081327
Antagonist Xist and Tsix co-transcription during mouse oogenesis and maternal Xist expression during pre-implantation development calls into question the nature of the maternal imprint on the X chromosome
Abstract
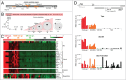
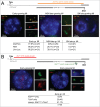
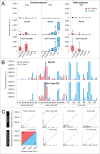
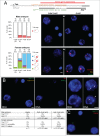
During the first divisions of the female mouse embryo, the paternal X-chromosome is coated by Xist non-coding RNA and gradually silenced. This imprinted X-inactivation principally results from the apposition, during oocyte growth, of an imprint on the X-inactivation master control region: the X-inactivation center (Xic). This maternal imprint of yet unknown nature is thought to prevent Xist upregulation from the maternal X (X(M)) during early female development. In order to provide further insight into the X(M) imprinting mechanism, we applied single-cell approaches to oocytes and pre-implantation embryos at different stages of development to analyze the expression of candidate genes within the Xic. We show that, unlike the situation pertaining in most other cellular contexts, in early-growing oocytes, Xist and Tsix sense and antisense transcription occur simultaneously from the same chromosome. Additionally, during early development, Xist appears to be transiently transcribed from the X(M) in some blastomeres of late 2-cell embryos concomitant with the general activation of the genome indicating that X(M) imprinting does not completely suppress maternal Xist transcription during embryo cleavage stages. These unexpected transcriptional regulations of the Xist locus call for a re-evaluation of the early functioning of the maternal imprint on the X-chromosome and suggest that Xist/Tsix antagonist transcriptional activities may participate in imprinting the maternal locus as described at other loci subject to parental imprinting.
Keywords: X-inactivation; imprinting; long non-coding RNAs; mouse oogenesis; mouse pre-implantation development; single-cell analysis; transcription.
Figures
Similar articles
-
Maintenance of Xist Imprinting Depends on Chromatin Condensation State and Rnf12 Dosage in Mice.PLoS Genet. 2016 Oct 27;12(10):e1006375. doi: 10.1371/journal.pgen.1006375. eCollection 2016 Oct. PLoS Genet. 2016. PMID: 27788132 Free PMC article.
-
A new Xist allele driven by a constitutively active promoter is dominated by Xist locus environment and exhibits the parent-of-origin effects.Development. 2015 Dec 15;142(24):4299-308. doi: 10.1242/dev.128819. Epub 2015 Oct 28. Development. 2015. PMID: 26511926
-
Genomic imprinting of Xist by maternal H3K27me3.Genes Dev. 2017 Oct 1;31(19):1927-1932. doi: 10.1101/gad.304113.117. Genes Dev. 2017. PMID: 29089420 Free PMC article.
-
What makes the maternal X chromosome resistant to undergoing imprinted X inactivation?Philos Trans R Soc Lond B Biol Sci. 2017 Nov 5;372(1733):20160365. doi: 10.1098/rstb.2016.0365. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28947661 Free PMC article. Review.
-
Dynamic interplay and function of multiple noncoding genes governing X chromosome inactivation.Biochim Biophys Acta. 2016 Jan;1859(1):112-20. doi: 10.1016/j.bbagrm.2015.07.015. Epub 2015 Aug 7. Biochim Biophys Acta. 2016. PMID: 26260844 Free PMC article. Review.
Cited by
-
Mutually exclusive sense-antisense transcription at FLC facilitates environmentally induced gene repression.Nat Commun. 2016 Oct 7;7:13031. doi: 10.1038/ncomms13031. Nat Commun. 2016. PMID: 27713408 Free PMC article.
-
DNA methylation and functional characterization of the XIST gene during in vitro early embryo development in cattle.Epigenetics. 2019 Jun;14(6):568-588. doi: 10.1080/15592294.2019.1600828. Epub 2019 Apr 12. Epigenetics. 2019. PMID: 30925851 Free PMC article.
References
-
- Kay GF, Barton SC, Surani MA, Rastan S. Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell 1994; 77:639-50; PMID:8205614; http://dx.doi.org/10.1016/0092-8674(94)90049-3 - DOI - PubMed
-
- Matsui J, Goto Y, Takagi N. Control of Xist expression for imprinted and random X chromosome inactivation in mice. Hum Mol Genet 2001; 10:1393-401; PMID:11440992; http://dx.doi.org/10.1093/hmg/10.13.1393 - DOI - PubMed
-
- Zuccotti M, Boiani M, Ponce R, Guizzardi S, Scandroglio R, Garagna S, Redi CA. Mouse Xist expression begins at zygotic genome activation and is timed by a zygotic clock. Mol Reprod Dev 2002; 61:14-20; PMID:11774371; http://dx.doi.org/10.1002/mrd.1126 - DOI - PubMed
-
- Nesterova TB, Barton SC, Surani MA, Brockdorff N. Loss of Xist imprinting in diploid parthenogenetic preimplantation embryos. Dev Biol 2001; 235:343-50; PMID:11437441; http://dx.doi.org/10.1006/dbio.2001.0295 - DOI - PubMed
-
- Okamoto I, Tan S, Takagi N. X-chromosome inactivation in XX androgenetic mouse embryos surviving implantation. Development 2000; 127:4137-45; PMID:10976046 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
