Mapping the Dynamics Landscape of Conformational Transitions in Enzyme: The Adenylate Kinase Case
- PMID: 26244746
- PMCID: PMC4572606
- DOI: 10.1016/j.bpj.2015.06.059
Mapping the Dynamics Landscape of Conformational Transitions in Enzyme: The Adenylate Kinase Case
Abstract
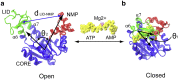
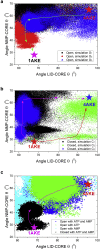
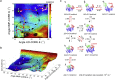
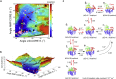
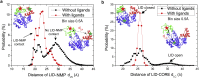
Conformational transition describes the essential dynamics and mechanism of enzymes in pursuing their various functions. The fundamental and practical challenge to researchers is to quantitatively describe the roles of large-scale dynamic transitions for regulating the catalytic processes. In this study, we tackled this challenge by exploring the pathways and free energy landscape of conformational changes in adenylate kinase (AdK), a key ubiquitous enzyme for cellular energy homeostasis. Using explicit long-timescale (up to microseconds) molecular dynamics and bias-exchange metadynamics simulations, we determined at the atomistic level the intermediate conformational states and mapped the transition pathways of AdK in the presence and absence of ligands. There is clearly chronological operation of the functional domains of AdK. Specifically in the ligand-free AdK, there is no significant energy barrier in the free energy landscape separating the open and closed states. Instead there are multiple intermediate conformational states, which facilitate the rapid transitions of AdK. In the ligand-bound AdK, the closed conformation is energetically most favored with a large energy barrier to open it up, and the conformational population prefers to shift to the closed form coupled with transitions. The results suggest a perspective for a hybrid of conformational selection and induced fit operations of ligand binding to AdK. These observations, depicted in the most comprehensive and quantitative way to date, to our knowledge, emphasize the underlying intrinsic dynamics of AdK and reveal the sophisticated conformational transitions of AdK in fulfilling its enzymatic functions. The developed methodology can also apply to other proteins and biomolecular systems.
Copyright © 2015 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Large-scale allosteric conformational transitions of adenylate kinase appear to involve a population-shift mechanism.Proc Natl Acad Sci U S A. 2007 Nov 20;104(47):18496-501. doi: 10.1073/pnas.0706443104. Epub 2007 Nov 13. Proc Natl Acad Sci U S A. 2007. PMID: 18000050 Free PMC article.
-
Exploring transition pathway and free-energy profile of large-scale protein conformational change by combining normal mode analysis and umbrella sampling molecular dynamics.J Phys Chem B. 2014 Jan 9;118(1):134-43. doi: 10.1021/jp4105129. Epub 2013 Dec 26. J Phys Chem B. 2014. PMID: 24350625
-
Modulation of the Conformational Dynamics of Apo-Adenylate Kinase through a π-Cation Interaction.J Phys Chem B. 2017 Jun 15;121(23):5699-5708. doi: 10.1021/acs.jpcb.7b01736. Epub 2017 Jun 6. J Phys Chem B. 2017. PMID: 28534408
-
Folding funnels and conformational transitions via hinge-bending motions.Cell Biochem Biophys. 1999;31(2):141-64. doi: 10.1007/BF02738169. Cell Biochem Biophys. 1999. PMID: 10593256 Review.
-
Markov state models provide insights into dynamic modulation of protein function.Acc Chem Res. 2015 Feb 17;48(2):414-22. doi: 10.1021/ar5002999. Epub 2015 Jan 3. Acc Chem Res. 2015. PMID: 25625937 Free PMC article. Review.
Cited by
-
Conformational dynamics of adenylate kinase in crystals.Struct Dyn. 2024 Feb 21;11(1):014702. doi: 10.1063/4.0000205. eCollection 2024 Jan. Struct Dyn. 2024. PMID: 38389978 Free PMC article.
-
Elucidating Dynamics of Adenylate Kinase from Enzyme Opening to Ligand Release.J Chem Inf Model. 2024 Jan 8;64(1):150-163. doi: 10.1021/acs.jcim.3c01618. Epub 2023 Dec 20. J Chem Inf Model. 2024. PMID: 38117131 Free PMC article.
-
Tracking the ATP-binding response in adenylate kinase in real time.Sci Adv. 2021 Nov 19;7(47):eabi5514. doi: 10.1126/sciadv.abi5514. Epub 2021 Nov 17. Sci Adv. 2021. PMID: 34788091 Free PMC article.
-
Role of Repeated Conformational Transitions in Substrate Binding of Adenylate Kinase.J Phys Chem B. 2022 Oct 20;126(41):8188-8201. doi: 10.1021/acs.jpcb.2c05497. Epub 2022 Oct 12. J Phys Chem B. 2022. PMID: 36222098 Free PMC article.
-
Tackling Hysteresis in Conformational Sampling: How to Be Forgetful with MEMENTO.J Chem Theory Comput. 2023 Jun 27;19(12):3705-3720. doi: 10.1021/acs.jctc.3c00140. Epub 2023 Jun 7. J Chem Theory Comput. 2023. PMID: 37285481 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

