Mediator independently orchestrates multiple steps of preinitiation complex assembly in vivo
- PMID: 26240385
- PMCID: PMC4627066
- DOI: 10.1093/nar/gkv782
Mediator independently orchestrates multiple steps of preinitiation complex assembly in vivo
Abstract
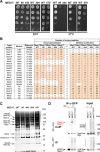
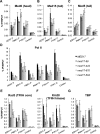
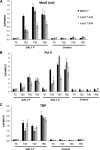
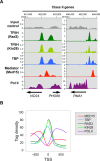
Mediator is a large multiprotein complex conserved in all eukaryotes, which has a crucial coregulator function in transcription by RNA polymerase II (Pol II). However, the molecular mechanisms of its action in vivo remain to be understood. Med17 is an essential and central component of the Mediator head module. In this work, we utilised our large collection of conditional temperature-sensitive med17 mutants to investigate Mediator's role in coordinating preinitiation complex (PIC) formation in vivo at the genome level after a transfer to a non-permissive temperature for 45 minutes. The effect of a yeast mutation proposed to be equivalent to the human Med17-L371P responsible for infantile cerebral atrophy was also analyzed. The ChIP-seq results demonstrate that med17 mutations differentially affected the global presence of several PIC components including Mediator, TBP, TFIIH modules and Pol II. Our data show that Mediator stabilizes TFIIK kinase and TFIIH core modules independently, suggesting that the recruitment or the stability of TFIIH modules is regulated independently on yeast genome. We demonstrate that Mediator selectively contributes to TBP recruitment or stabilization to chromatin. This study provides an extensive genome-wide view of Mediator's role in PIC formation, suggesting that Mediator coordinates multiple steps of a PIC assembly pathway.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Toward understanding of the mechanisms of Mediator function in vivo: Focus on the preinitiation complex assembly.Transcription. 2017;8(5):328-342. doi: 10.1080/21541264.2017.1329000. Epub 2017 Aug 25. Transcription. 2017. PMID: 28841352 Free PMC article. Review.
-
Mediator-dependent recruitment of TFIIH modules in preinitiation complex.Mol Cell. 2008 Aug 8;31(3):337-46. doi: 10.1016/j.molcel.2008.06.021. Mol Cell. 2008. PMID: 18691966
-
Direct interaction of RNA polymerase II and mediator required for transcription in vivo.Science. 2011 Mar 18;331(6023):1451-4. doi: 10.1126/science.1200188. Science. 2011. PMID: 21415355
-
Functional interplay between Mediator and TFIIB in preinitiation complex assembly in relation to promoter architecture.Genes Dev. 2016 Sep 15;30(18):2119-2132. doi: 10.1101/gad.285775.116. Epub 2016 Sep 29. Genes Dev. 2016. PMID: 27688401 Free PMC article.
-
More pieces to the puzzle: recent structural insights into class II transcription initiation.Curr Opin Struct Biol. 2014 Feb;24:91-7. doi: 10.1016/j.sbi.2013.12.005. Epub 2014 Jan 16. Curr Opin Struct Biol. 2014. PMID: 24440461 Review.
Cited by
-
Role of the pre-initiation complex in Mediator recruitment and dynamics.Elife. 2018 Dec 12;7:e39633. doi: 10.7554/eLife.39633. Elife. 2018. PMID: 30540252 Free PMC article.
-
SAGA Is a General Cofactor for RNA Polymerase II Transcription.Mol Cell. 2017 Oct 5;68(1):130-143.e5. doi: 10.1016/j.molcel.2017.08.016. Epub 2017 Sep 14. Mol Cell. 2017. PMID: 28918903 Free PMC article.
-
A Role for the Mre11-Rad50-Xrs2 Complex in Gene Expression and Chromosome Organization.Mol Cell. 2021 Jan 7;81(1):183-197.e6. doi: 10.1016/j.molcel.2020.11.010. Epub 2020 Dec 4. Mol Cell. 2021. PMID: 33278361 Free PMC article.
-
Toward understanding of the mechanisms of Mediator function in vivo: Focus on the preinitiation complex assembly.Transcription. 2017;8(5):328-342. doi: 10.1080/21541264.2017.1329000. Epub 2017 Aug 25. Transcription. 2017. PMID: 28841352 Free PMC article. Review.
-
Functional interplay between Mediator and RNA polymerase II in Rad2/XPG loading to the chromatin.Nucleic Acids Res. 2019 Sep 26;47(17):8988-9004. doi: 10.1093/nar/gkz598. Nucleic Acids Res. 2019. PMID: 31299084 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

