In-depth analysis of the critical genes and pathways in colorectal cancer
- PMID: 26239303
- PMCID: PMC4564077
- DOI: 10.3892/ijmm.2015.2298
In-depth analysis of the critical genes and pathways in colorectal cancer
Abstract
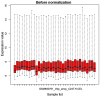
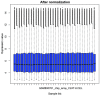
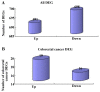
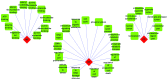
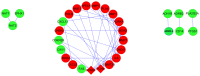
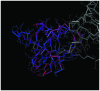
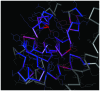
The present study aimed to investigate the molecular targets for colorectal cancer (CRC). Differentially expressed genes (DEGs) were screened between CRC and matched adjacent noncancerous samples. GENETIC_ASSOIATION_DB_DISEASE analysis was performed to identify CRC genes from the identified DEGs using the Database for Annotation, Visualization and Integrated Discovery, followed by Gene Οntology (GO) and Kyoto Encyclopedia of Genes and Genomes analysis for the CRC genes. A protein‑protein interaction (PPI) network was constructed for the CRC genes, followed by determination and analysis of the hub genes, in terms of the protein domains and spatial structure. In total, 35 CRC genes were determined, including 19 upregulated and 16 downregulated genes. Downregulated N‑acetyltransferase (NAT)1 and NAT2 were enriched in the caffeine metabolism pathway. The downregulated and upregulated genes were enriched in a number of GO terms and pathways, respectively. Cyclin D1 (CCND1) and proliferating cell nuclear antigen (PCNA) were identified as the hub genes in the PPI network. The C‑terminal and N‑terminal domains were similar in PCNA, but different in CCND1. The results suggested PCNA, CCND1, NAT1 and NAT2 for use as biomarkers to enable early diagnosis and monitoring of CRC. These results form a basis for developing therapies, which target the unique protein domains of PCNA and CCND1.
Figures

Similar articles
-
Screening key genes and signaling pathways in colorectal cancer by integrated bioinformatics analysis.Mol Med Rep. 2019 Aug;20(2):1259-1269. doi: 10.3892/mmr.2019.10336. Epub 2019 Jun 4. Mol Med Rep. 2019. PMID: 31173250 Free PMC article.
-
Identification of key genes in colorectal cancer using random walk with restart.Mol Med Rep. 2017 Feb;15(2):867-872. doi: 10.3892/mmr.2016.6058. Epub 2016 Dec 19. Mol Med Rep. 2017. PMID: 28000901
-
Integrated analysis of differentially expressed genes and pathways in triple‑negative breast cancer.Mol Med Rep. 2017 Mar;15(3):1087-1094. doi: 10.3892/mmr.2017.6101. Epub 2017 Jan 4. Mol Med Rep. 2017. PMID: 28075450 Free PMC article.
-
Delineating the underlying molecular mechanisms and key genes involved in metastasis of colorectal cancer via bioinformatics analysis.Oncol Rep. 2018 May;39(5):2297-2305. doi: 10.3892/or.2018.6303. Epub 2018 Mar 8. Oncol Rep. 2018. PMID: 29517105
-
Identification of Key Genes in Colorectal Cancer Regulated by miR-34a.Med Sci Monit. 2017 Dec 3;23:5735-5743. doi: 10.12659/msm.904937. Med Sci Monit. 2017. PMID: 29197895 Free PMC article.
Cited by
-
Genome-wide identification of long non-coding RNA and mRNA profiling using RNA sequencing in subjects with sensitive skin.Oncotarget. 2017 Dec 12;8(70):114894-114910. doi: 10.18632/oncotarget.23147. eCollection 2017 Dec 29. Oncotarget. 2017. PMID: 29383128 Free PMC article.
-
Rapid preliminary purity evaluation of tumor biopsies using deep learning approach.Comput Struct Biotechnol J. 2020 Jun 16;18:1746-1753. doi: 10.1016/j.csbj.2020.06.007. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32695267 Free PMC article.
-
Novel diagnostic and prognostic biomarkers of colorectal cancer: Capable to overcome the heterogeneity-specific barrier and valid for global applications.PLoS One. 2021 Sep 2;16(9):e0256020. doi: 10.1371/journal.pone.0256020. eCollection 2021. PLoS One. 2021. PMID: 34473751 Free PMC article.
-
Identification of lymph node metastasis-related genes and patterns of immune infiltration in colon adenocarcinoma.Front Oncol. 2023 Jan 16;12:907464. doi: 10.3389/fonc.2022.907464. eCollection 2022. Front Oncol. 2023. PMID: 36727052 Free PMC article.
-
Supplementation with High or Low Iron Reduces Colitis Severity in an AOM/DSS Mouse Model.Nutrients. 2022 May 12;14(10):2033. doi: 10.3390/nu14102033. Nutrients. 2022. PMID: 35631174 Free PMC article.
References
-
- Stewart BW, Wild C. World Cancer Report 2014. World Health Organization; 2014.
-
- Stegeman I, de Wijkerslooth TR, Stoop EM, van Leerdam ME, Dekker E, van Ballegooijen M, Kuipers EJ, Fockens P, Kraaijenhagen RA, Bossuyt PM. Colorectal cancer risk factors in the detection of advanced adenoma and colorectal cancer. Cancer Epidemiol. 2013;37:278–283. doi: 10.1016/j.canep.2013.02.004. - DOI - PubMed
-
- Goyette MC, Cho K, Fasching CL, Levy DB, Kinzler KW, Paraskeva C, Vogelstein B, Stanbridge EJ. Progression of colorectal cancer is associated with multiple tumor suppressor gene defects but inhibition of tumorigenicity is accomplished by correction of any single defect via chromosome transfer. Mol Cell Biol. 1992;12:1387–1395. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

