The N-terminus of RPA large subunit and its spatial position are important for the 5'->3' resection of DNA double-strand breaks
- PMID: 26227969
- PMCID: PMC4605295
- DOI: 10.1093/nar/gkv764
The N-terminus of RPA large subunit and its spatial position are important for the 5'->3' resection of DNA double-strand breaks
Abstract
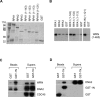
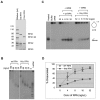
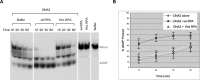
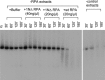
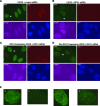
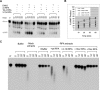
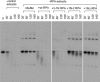
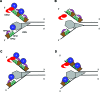
The first step of homology-dependent repair of DNA double-strand breaks (DSBs) is the resection of the 5' strand to generate 3' ss-DNA. Of the two major nucleases responsible for resection, EXO1 has intrinsic 5'->3' directionality, but DNA2 does not. DNA2 acts with RecQ helicases such as the Werner syndrome protein (WRN) and the heterotrimeric eukaryotic ss-DNA binding protein RPA. We have found that the N-terminus of the RPA large subunit (RPA1N) interacts with both WRN and DNA2 and is essential for stimulating WRN's 3'->5' helicase activity and DNA2's 5'->3' ss-DNA exonuclease activity. A mutant RPA complex that lacks RPA1N is unable to support resection in Xenopus egg extracts and human cells. Furthermore, relocating RPA1N to the middle subunit but not to the small subunit causes severe defects in stimulating DNA2 and WRN and in supporting resection. Together, these findings suggest that RPA1N and its spatial position are critical for restricting the directionality of the WRN-DNA2 resection pathway.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Replication protein A promotes 5'-->3' end processing during homology-dependent DNA double-strand break repair.J Cell Biol. 2011 Jan 24;192(2):251-61. doi: 10.1083/jcb.201005110. J Cell Biol. 2011. PMID: 21263027 Free PMC article.
-
Mechanistic analysis of Xenopus EXO1's function in 5'-strand resection at DNA double-strand breaks.Nucleic Acids Res. 2011 Aug;39(14):5967-77. doi: 10.1093/nar/gkr216. Epub 2011 Apr 13. Nucleic Acids Res. 2011. PMID: 21490081 Free PMC article.
-
Analysis of MRE11's function in the 5'-->3' processing of DNA double-strand breaks.Nucleic Acids Res. 2012 May;40(10):4496-506. doi: 10.1093/nar/gks044. Epub 2012 Feb 8. Nucleic Acids Res. 2012. PMID: 22319209 Free PMC article.
-
Werner syndrome protein: biochemical properties and functional interactions.Exp Gerontol. 2000 Sep;35(6-7):695-702. doi: 10.1016/s0531-5565(00)00145-5. Exp Gerontol. 2000. PMID: 11053659 Review.
-
The Werner syndrome gene: the molecular basis of RecQ helicase-deficiency diseases.Trends Genet. 2000 May;16(5):213-20. doi: 10.1016/s0168-9525(99)01970-8. Trends Genet. 2000. PMID: 10782115 Review.
Cited by
-
Distinct RPA domains promote recruitment and the helicase-nuclease activities of Dna2.Nat Commun. 2021 Nov 11;12(1):6521. doi: 10.1038/s41467-021-26863-y. Nat Commun. 2021. PMID: 34764291 Free PMC article.
-
Multiple RPAs make WRN syndrome protein a superhelicase.Nucleic Acids Res. 2018 May 18;46(9):4689-4698. doi: 10.1093/nar/gky272. Nucleic Acids Res. 2018. PMID: 29668972 Free PMC article.
-
CDK1 phosphorylates WRN at collapsed replication forks.Nat Commun. 2016 Sep 16;7:12880. doi: 10.1038/ncomms12880. Nat Commun. 2016. PMID: 27634057 Free PMC article.
-
Investigation of the core binding regions of human Werner syndrome and Fanconi anemia group J helicases on replication protein A.Sci Rep. 2019 Sep 30;9(1):14016. doi: 10.1038/s41598-019-50502-8. Sci Rep. 2019. PMID: 31570747 Free PMC article.
-
Sharpening the ends for repair: mechanisms and regulation of DNA resection.Acta Biochim Biophys Sin (Shanghai). 2016 Jul;48(7):647-57. doi: 10.1093/abbs/gmw043. Epub 2016 May 12. Acta Biochim Biophys Sin (Shanghai). 2016. PMID: 27174871 Free PMC article. Review.
References
-
- Nussenzweig A., Nussenzweig M.C. A backup DNA repair pathway moves to the forefront. Cell. 2007;131:223–225. - PubMed
-
- Symington L.S., Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. - PubMed
-
- Toczylowski T., Yan H. Mechanistic analysis of a DNA end processing pathway mediated by the Xenopus Werner syndrome protein. J. Biol. Chem. 2006;281:33198–33205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

