Protein Folding and Mechanisms of Proteostasis
- PMID: 26225966
- PMCID: PMC4581189
- DOI: 10.3390/ijms160817193
Protein Folding and Mechanisms of Proteostasis
Abstract
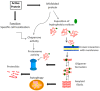
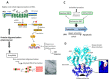
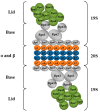
Highly sophisticated mechanisms that modulate protein structure and function, which involve synthesis and degradation, have evolved to maintain cellular homeostasis. Perturbations in these mechanisms can lead to protein dysfunction as well as deleterious cell processes. Therefore in recent years the etiology of a great number of diseases has been attributed to failures in mechanisms that modulate protein structure. Interconnections among metabolic and cell signaling pathways are critical for homeostasis to converge on mechanisms associated with protein folding as well as for the preservation of the native structure of proteins. For instance, imbalances in secretory protein synthesis pathways lead to a condition known as endoplasmic reticulum (ER) stress which elicits the adaptive unfolded protein response (UPR). Therefore, taking this into consideration, a key part of this paper is developed around the protein folding phenomenon, and cellular mechanisms which support this pivotal condition. We provide an overview of chaperone protein function, UPR via, spatial compartmentalization of protein folding, proteasome role, autophagy, as well as the intertwining between these processes. Several diseases are known to have a molecular etiology in the malfunction of mechanisms responsible for protein folding and in the shielding of native structure, phenomena which ultimately lead to misfolded protein accumulation. This review centers on our current knowledge about pathways that modulate protein folding, and cell responses involved in protein homeostasis.
Keywords: folding; misfolding; proteins; proteostasis.
Figures

Similar articles
-
Molecular Chaperones in Neurodegenerative Diseases: A Short Review.Adv Exp Med Biol. 2017;987:219-231. doi: 10.1007/978-3-319-57379-3_20. Adv Exp Med Biol. 2017. PMID: 28971461 Review.
-
The disturbance of protein synthesis/degradation homeostasis is a common trait of age-related neurodegenerative disorders.Adv Protein Chem Struct Biol. 2022;132:49-87. doi: 10.1016/bs.apcsb.2022.05.008. Epub 2022 Jun 9. Adv Protein Chem Struct Biol. 2022. PMID: 36088079
-
The unfolded protein response: a stress signaling pathway critical for health and disease.Neurology. 2006 Jan 24;66(2 Suppl 1):S102-9. doi: 10.1212/01.wnl.0000192306.98198.ec. Neurology. 2006. PMID: 16432136 Review.
-
ER and aging-Protein folding and the ER stress response.Ageing Res Rev. 2009 Jul;8(3):150-9. doi: 10.1016/j.arr.2009.03.001. Epub 2009 Mar 21. Ageing Res Rev. 2009. PMID: 19491040 Review.
-
Modeling the Critical Activation of Chaperone Machinery in Protein Folding.Adv Exp Med Biol. 2020;1194:351-358. doi: 10.1007/978-3-030-32622-7_33. Adv Exp Med Biol. 2020. PMID: 32468551
Cited by
-
Protective Roles of Cytosolic and Plastidal Proteasomes on Abiotic Stress and Pathogen Invasion.Plants (Basel). 2020 Jul 2;9(7):832. doi: 10.3390/plants9070832. Plants (Basel). 2020. PMID: 32630761 Free PMC article. Review.
-
Insulin Is a Key Modulator of Fetoplacental Endothelium Metabolic Disturbances in Gestational Diabetes Mellitus.Front Physiol. 2016 Mar 31;7:119. doi: 10.3389/fphys.2016.00119. eCollection 2016. Front Physiol. 2016. PMID: 27065887 Free PMC article. Review.
-
Radiovaccination Strategy for Cancer Treatment Integrating Photodynamic Therapy-Generated Vaccines with Radiotherapy.Int J Mol Sci. 2022 Oct 14;23(20):12263. doi: 10.3390/ijms232012263. Int J Mol Sci. 2022. PMID: 36293116 Free PMC article. Review.
-
78 kDa Glucose-Regulated Protein Attenuates Protein Aggregation and Monocyte Adhesion Induced by Angiotensin II in Vascular Cells.Int J Mol Sci. 2020 Jul 15;21(14):4980. doi: 10.3390/ijms21144980. Int J Mol Sci. 2020. PMID: 32679678 Free PMC article.
-
FOXR1 regulates stress response pathways and is necessary for proper brain development.PLoS Genet. 2021 Nov 1;17(11):e1009854. doi: 10.1371/journal.pgen.1009854. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34723967 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

