The Ku subunit of telomerase binds Sir4 to recruit telomerase to lengthen telomeres in S. cerevisiae
- PMID: 26218225
- PMCID: PMC4547093
- DOI: 10.7554/eLife.07750
The Ku subunit of telomerase binds Sir4 to recruit telomerase to lengthen telomeres in S. cerevisiae
Abstract
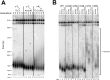
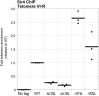
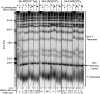
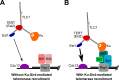
In Saccharomyces cerevisiae and in humans, the telomerase RNA subunit is bound by Ku, a ring-shaped protein heterodimer best known for its function in DNA repair. Ku binding to yeast telomerase RNA promotes telomere lengthening and telomerase recruitment to telomeres, but how this is achieved remains unknown. Using telomere-length analysis and chromatin immunoprecipitation, we show that Sir4 - a previously identified Ku-binding protein that is a component of telomeric silent chromatin - is required for Ku-mediated telomere lengthening and telomerase recruitment. We also find that specifically tethering Sir4 directly to Ku-binding-defective telomerase RNA restores otherwise-shortened telomeres to wild-type length. These findings suggest that Sir4 is the telomere-bound target of Ku-mediated telomerase recruitment and provide one mechanism for how the Sir4-competing Rif1 and Rif2 proteins negatively regulate telomere length in yeast.
Keywords: Ku; S. cerevisiae; chromosomes; genes; telomerase; telomere; transcriptional silencing; yeast.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures



Similar articles
-
Ku must load directly onto the chromosome end in order to mediate its telomeric functions.PLoS Genet. 2011 Aug;7(8):e1002233. doi: 10.1371/journal.pgen.1002233. Epub 2011 Aug 11. PLoS Genet. 2011. PMID: 21852961 Free PMC article.
-
Structural Insights into Yeast Telomerase Recruitment to Telomeres.Cell. 2018 Jan 11;172(1-2):331-343.e13. doi: 10.1016/j.cell.2017.12.008. Epub 2017 Dec 28. Cell. 2018. PMID: 29290466 Free PMC article.
-
Cell cycle-dependent regulation of yeast telomerase by Ku.Nat Struct Mol Biol. 2004 Dec;11(12):1198-205. doi: 10.1038/nsmb854. Epub 2004 Nov 7. Nat Struct Mol Biol. 2004. PMID: 15531893
-
Regulation of telomerase by telomeric proteins.Annu Rev Biochem. 2004;73:177-208. doi: 10.1146/annurev.biochem.73.071403.160049. Annu Rev Biochem. 2004. PMID: 15189140 Review.
-
Telomeres--unsticky ends.Science. 1998 Sep 18;281(5384):1818-9. doi: 10.1126/science.281.5384.1818. Science. 1998. PMID: 9776685 Review. No abstract available.
Cited by
-
Telomerase, the recombination machinery and Rap1 play redundant roles in yeast telomere protection.Curr Genet. 2021 Feb;67(1):153-163. doi: 10.1007/s00294-020-01125-4. Epub 2020 Nov 6. Curr Genet. 2021. PMID: 33156376
-
Telomerase structures and regulation: shedding light on the chromosome end.Curr Opin Struct Biol. 2019 Apr;55:185-193. doi: 10.1016/j.sbi.2019.04.009. Epub 2019 Jun 12. Curr Opin Struct Biol. 2019. PMID: 31202023 Free PMC article. Review.
-
Repositioning the Sm-Binding Site in Saccharomyces cerevisiae Telomerase RNA Reveals RNP Organizational Flexibility and Sm-Directed 3'-End Formation.Noncoding RNA. 2020 Feb 29;6(1):9. doi: 10.3390/ncrna6010009. Noncoding RNA. 2020. PMID: 32121425 Free PMC article.
-
Functional characterisation of long intergenic non-coding RNAs through genetic interaction profiling in Saccharomyces cerevisiae.BMC Biol. 2016 Dec 7;14(1):106. doi: 10.1186/s12915-016-0325-7. BMC Biol. 2016. PMID: 27927215 Free PMC article.
-
Telomere length regulation by Rif1 protein from Hansenula polymorpha.Elife. 2022 Feb 7;11:e75010. doi: 10.7554/eLife.75010. Elife. 2022. PMID: 35129114 Free PMC article.
References
-
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proceedings of the National Academy of Sciences of USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

