Gene-expression analysis of a colorectal cancer-specific discriminatory transcript set on formalin-fixed, paraffin-embedded (FFPE) tissue samples
- PMID: 26208990
- PMCID: PMC4515026
- DOI: 10.1186/s13000-015-0363-4
Gene-expression analysis of a colorectal cancer-specific discriminatory transcript set on formalin-fixed, paraffin-embedded (FFPE) tissue samples
Abstract
Background: A recently published transcript set is suitable for gene expression-based discrimination of normal colonic and colorectal cancer (CRC) biopsy samples. Our aim was to test the discriminatory power of the CRC-specific transcript set on independent biopsies and on formalin-fixed, paraffin-embedded (FFPE) tissue samples.
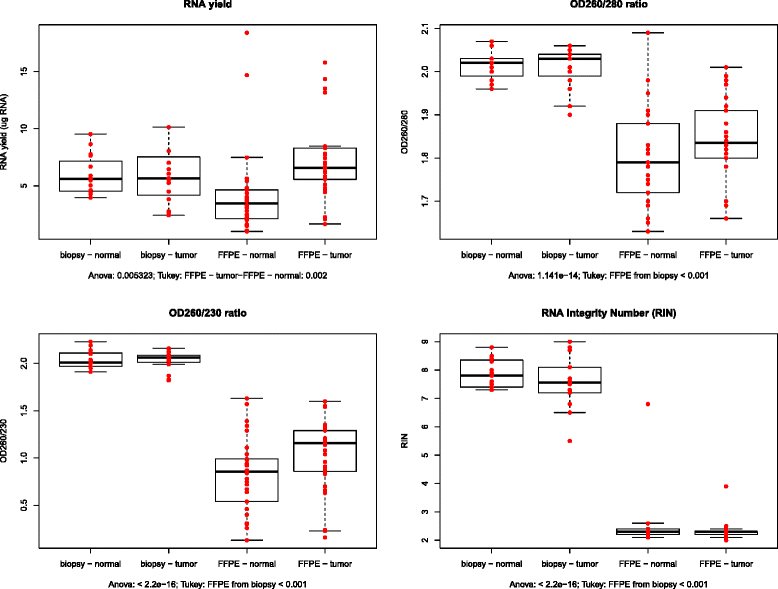
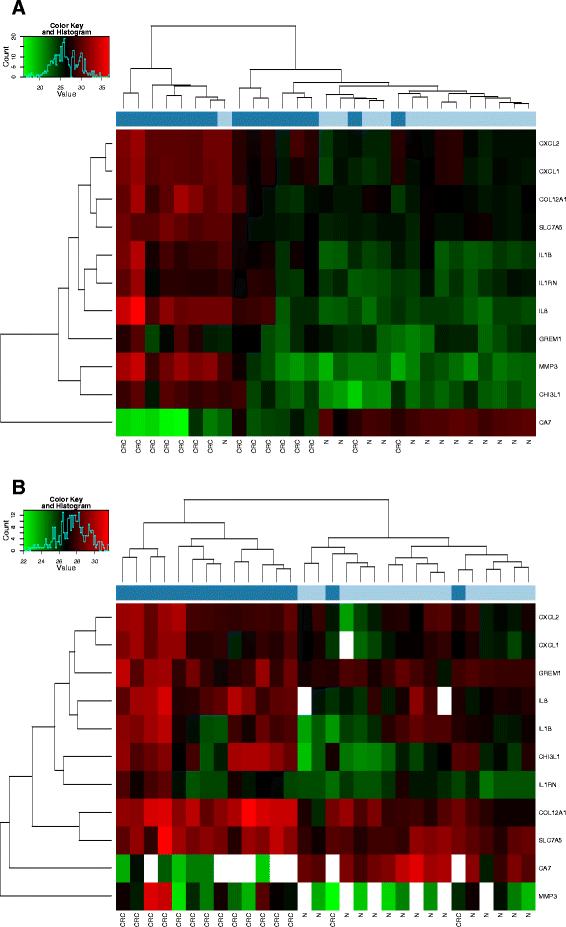
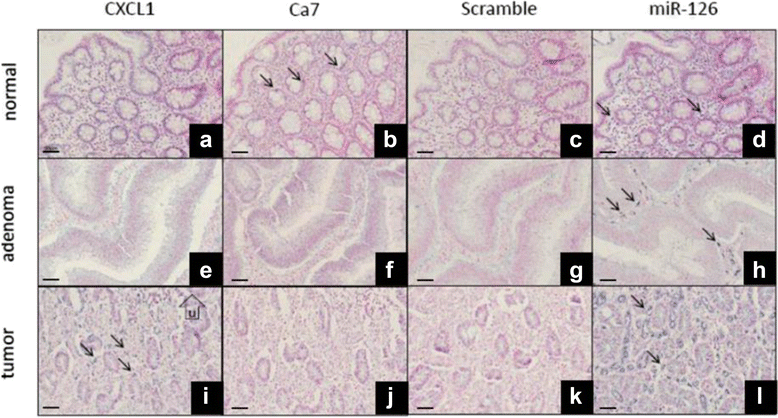
Methods: Total RNA isolations were performed with the automated MagNA Pure 96 Cellular RNA Large Volume Kit (Roche) from fresh frozen biopsies stored in RNALater (CRC (n = 15) and healthy colonic (n = 15)), furthermore from FFPE specimens including CRC (n = 15) and normal adjacent tissue (NAT) (n = 15) specimens next to the tumor. After quality and quantity measurements, gene expression analysis of a colorectal cancer-specific marker set with 11 genes (CA7, COL12A1, CXCL1, CXCL2, CHI3L1, GREM1, IL1B, IL1RN, IL8, MMP3, SLC5A7) was performed with array real-time PCR using Transcriptor First Strand cDNA Synthesis Kit (Roche) and RealTime ready assays on LightCycler480 System (Roche). In situ hybridization for two selected transcripts (CA7, CXCL1) was performed on NAT (n = 3), adenoma (n = 3) and CRC (n = 3) FFPE samples.
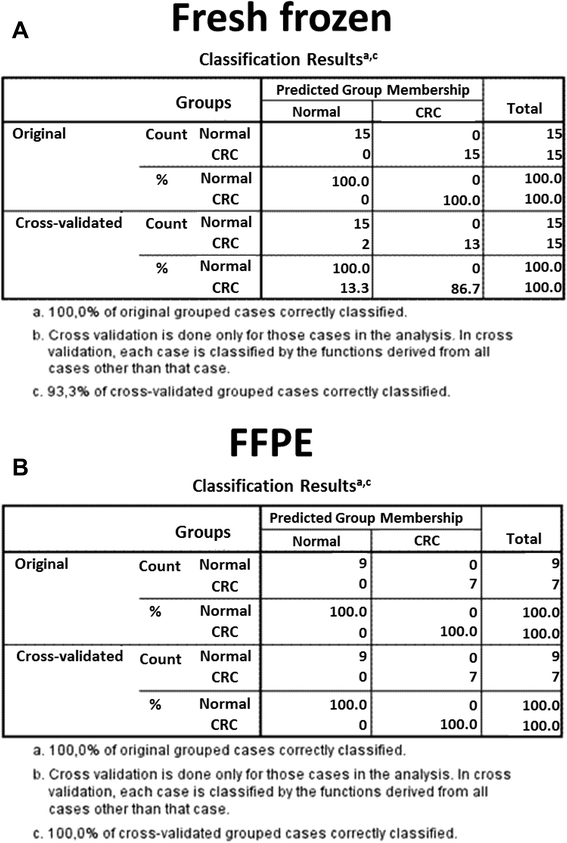
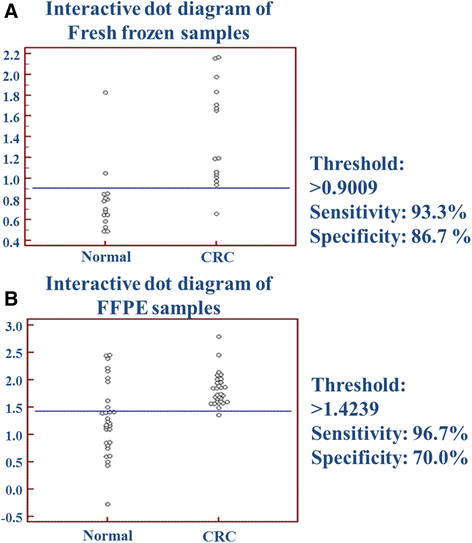
Results: Although analytical parameters of automatically isolated RNA samples showed differences between fresh frozen biopsy and FFPE samples, both quantity and the quality enabled their application in gene expression analyses. CRC and normal fresh frozen biopsy samples could be distinguished with 93.3% sensitivity and 86.7% specificity and FFPE samples with 96.7 and 70.0%, respectively. In situ hybridization could confirm the upregulation of CXCL1 and downregulation of CA7 in colorectal adenomas and tumors compared to healthy controls.
Conclusion: According to our results, gene expression analysis of the analyzed colorectal cancer-specific marker set can also be performed from FFPE tissue material. With the addition of an automated workflow, this marker set may enhance the objective classification of colorectal neoplasias in the routine procedure in the future.
Figures
Similar articles
-
Gene expression analysis of normal and colorectal cancer tissue samples from fresh frozen and matched formalin-fixed, paraffin-embedded (FFPE) specimens after manual and automated RNA isolation.Methods. 2013 Jan;59(1):S16-9. doi: 10.1016/j.ymeth.2012.09.011. Epub 2012 Oct 2. Methods. 2013. PMID: 23036325
-
A formalin-fixed paraffin-embedded (FFPE)-based prognostic signature to predict metastasis in clinically low risk stage I/II microsatellite stable colorectal cancer.Cancer Lett. 2017 Sep 10;403:13-20. doi: 10.1016/j.canlet.2017.05.031. Epub 2017 Jun 15. Cancer Lett. 2017. PMID: 28624625
-
Comparison of Automated and Manual DNA Isolation Methods for DNA Methylation Analysis of Biopsy, Fresh Frozen, and Formalin-Fixed, Paraffin-Embedded Colorectal Cancer Samples.J Lab Autom. 2015 Dec;20(6):642-51. doi: 10.1177/2211068214565903. Epub 2015 Jan 9. J Lab Autom. 2015. PMID: 25576093
-
Fusion transcript discovery using RNA sequencing in formalin-fixed paraffin-embedded specimen.Crit Rev Oncol Hematol. 2021 Apr;160:103303. doi: 10.1016/j.critrevonc.2021.103303. Epub 2021 Mar 20. Crit Rev Oncol Hematol. 2021. PMID: 33757837 Review.
-
Accuracy of Molecular Data Generated with FFPE Biospecimens: Lessons from the Literature.Cancer Res. 2015 Apr 15;75(8):1541-7. doi: 10.1158/0008-5472.CAN-14-2378. Epub 2015 Apr 2. Cancer Res. 2015. PMID: 25836717 Free PMC article. Review.
Cited by
-
Robust transcriptional tumor signatures applicable to both formalin-fixed paraffin-embedded and fresh-frozen samples.Oncotarget. 2017 Jan 24;8(4):6652-6662. doi: 10.18632/oncotarget.14257. Oncotarget. 2017. PMID: 28036264 Free PMC article.
-
Risk analysis of colorectal cancer incidence by gene expression analysis.PeerJ. 2017 Feb 15;5:e3003. doi: 10.7717/peerj.3003. eCollection 2017. PeerJ. 2017. PMID: 28229027 Free PMC article.
-
Novel circulating microRNAs expression profile in colon cancer: a pilot study.Eur J Med Res. 2017 Nov 29;22(1):51. doi: 10.1186/s40001-017-0294-5. Eur J Med Res. 2017. PMID: 29187262 Free PMC article.
-
Genetic Signatures From RNA Sequencing of Pediatric Localized Scleroderma Skin.Front Pediatr. 2021 Jun 7;9:669116. doi: 10.3389/fped.2021.669116. eCollection 2021. Front Pediatr. 2021. PMID: 34164359 Free PMC article.
References
-
- Huerta S. Recent advances in the molecular diagnosis and prognosis of colorectal cancer. Expert review of molecular diagnostics. 2008;8:277–88. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

