Helicase processivity and not the unwinding velocity exhibits universal increase with force
- PMID: 26200858
- PMCID: PMC4621497
- DOI: 10.1016/j.bpj.2015.05.020
Helicase processivity and not the unwinding velocity exhibits universal increase with force
Abstract
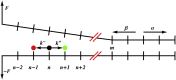
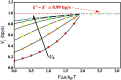
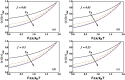
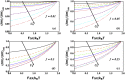
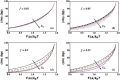
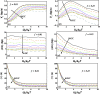
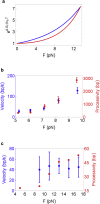
Helicases, involved in a number of cellular functions, are motors that translocate along single-stranded nucleic acid and couple the motion to unwinding double-strands of a duplex nucleic acid. The junction between double- and single-strands creates a barrier to the movement of the helicase, which can be manipulated in vitro by applying mechanical forces directly on the nucleic acid strands. Single-molecule experiments have demonstrated that the unwinding velocities of some helicases increase dramatically with increase in the external force, while others show little response. In contrast, the unwinding processivity always increases when the force increases. The differing responses of the unwinding velocity and processivity to force have lacked explanation. By generalizing a previous model of processive unwinding by helicases, we provide a unified framework for understanding the dependence of velocity and processivity on force and the nucleic acid sequence. We predict that the sensitivity of unwinding processivity to external force is a universal feature that should be observed in all helicases. Our prediction is illustrated using T7 and NS3 helicases as case studies. Interestingly, the increase in unwinding processivity with force depends on whether the helicase forces basepair opening by direct interaction or if such a disruption occurs spontaneously due to thermal fluctuations. Based on the theoretical results, we propose that proteins like single-strand binding proteins associated with helicases in the replisome may have coevolved with helicases to increase the unwinding processivity even if the velocity remains unaffected.
Copyright © 2015 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Processivity, Velocity, and Universal Characteristics of Nucleic Acid Unwinding by Helicases.Biophys J. 2019 Sep 3;117(5):867-879. doi: 10.1016/j.bpj.2019.07.021. Epub 2019 Jul 20. Biophys J. 2019. PMID: 31400912 Free PMC article.
-
Processivity of nucleic acid unwinding and translocation by helicases.Proteins. 2016 Nov;84(11):1590-1605. doi: 10.1002/prot.25102. Epub 2016 Jul 22. Proteins. 2016. PMID: 27410462
-
Dynamics of DNA unwinding by helicases with frequent backward steps.Math Biosci. 2017 Dec;294:33-45. doi: 10.1016/j.mbs.2017.10.004. Epub 2017 Oct 10. Math Biosci. 2017. PMID: 29024746
-
A mechanistic study of helicases with magnetic traps.Protein Sci. 2017 Jul;26(7):1314-1336. doi: 10.1002/pro.3187. Epub 2017 Jun 13. Protein Sci. 2017. PMID: 28474797 Free PMC article. Review.
-
Structure and function of hexameric helicases.Annu Rev Biochem. 2000;69:651-97. doi: 10.1146/annurev.biochem.69.1.651. Annu Rev Biochem. 2000. PMID: 10966472 Review.
Cited by
-
Human HELB is a processive motor protein that catalyzes RPA clearance from single-stranded DNA.Proc Natl Acad Sci U S A. 2022 Apr 12;119(15):e2112376119. doi: 10.1073/pnas.2112376119. Epub 2022 Apr 6. Proc Natl Acad Sci U S A. 2022. PMID: 35385349 Free PMC article.
-
Regulation of Rep helicase unwinding by an auto-inhibitory subdomain.Nucleic Acids Res. 2019 Mar 18;47(5):2523-2532. doi: 10.1093/nar/gkz023. Nucleic Acids Res. 2019. PMID: 30690484 Free PMC article.
-
Processivity, Velocity, and Universal Characteristics of Nucleic Acid Unwinding by Helicases.Biophys J. 2019 Sep 3;117(5):867-879. doi: 10.1016/j.bpj.2019.07.021. Epub 2019 Jul 20. Biophys J. 2019. PMID: 31400912 Free PMC article.
-
Single molecule kinetics uncover roles for E. coli RecQ DNA helicase domains and interaction with SSB.Nucleic Acids Res. 2018 Sep 19;46(16):8500-8515. doi: 10.1093/nar/gky647. Nucleic Acids Res. 2018. PMID: 30053104 Free PMC article.
-
Pif1 is a force-regulated helicase.Nucleic Acids Res. 2016 May 19;44(9):4330-9. doi: 10.1093/nar/gkw295. Epub 2016 Apr 20. Nucleic Acids Res. 2016. PMID: 27098034 Free PMC article.
References
-
- Lohman T.M. Escherichia coli DNA helicases: mechanisms of DNA unwinding. Mol. Microbiol. 1992;6:5–14. - PubMed
-
- Lohman T.M., Bjornson K.P. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. - PubMed
-
- Delagoutte E., von Hippel P.H. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I. Structures and properties of isolated helicases. Q. Rev. Biophys. 2002;35:431–478. - PubMed
-
- Delagoutte E., von Hippel P.H. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II. Integration of helicases into cellular processes. Q. Rev. Biophys. 2003;36:1–69. - PubMed
-
- Bianco P.R., Kowalczykowski S.C. Translocation step size and mechanism of the RecBC DNA helicase. Nature. 2000;405:368–372. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

