Rapid Identification of Chemoresistance Mechanisms Using Yeast DNA Mismatch Repair Mutants
- PMID: 26199284
- PMCID: PMC4555229
- DOI: 10.1534/g3.115.020560
Rapid Identification of Chemoresistance Mechanisms Using Yeast DNA Mismatch Repair Mutants
Abstract
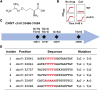
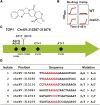
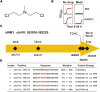
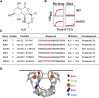
Resistance to cancer therapy is a major obstacle in the long-term treatment of cancer. A greater understanding of drug resistance mechanisms will ultimately lead to the development of effective therapeutic strategies to prevent resistance from occurring. Here, we exploit the mutator phenotype of mismatch repair defective yeast cells combined with whole genome sequencing to identify drug resistance mutations in key pathways involved in the development of chemoresistance. The utility of this approach was demonstrated via the identification of the known CAN1 and TOP1 resistance targets for two compounds, canavanine and camptothecin, respectively. We have also experimentally validated the plasma membrane transporter HNM1 as the primary drug resistance target of mechlorethamine. Furthermore, the sequencing of mitoxantrone-resistant strains identified inactivating mutations within IPT1, a gene encoding inositolphosphotransferase, an enzyme involved in sphingolipid biosynthesis. In the case of bactobolin, a promising anticancer drug, the endocytosis pathway was identified as the drug resistance target responsible for conferring resistance. Finally, we show that that rapamycin, an mTOR inhibitor previously shown to alter the fitness of the ipt1 mutant, can effectively prevent the formation of mitoxantrone resistance. The rapid and robust nature of these techniques, using Saccharomyces cerevisiae as a model organism, should accelerate the identification of drug resistance targets and guide the development of novel therapeutic combination strategies to prevent the development of chemoresistance in various cancers.
Keywords: DNA mismatch repair; cancer; drug resistance; mutator; whole genome sequencing.
Copyright © 2015 Ojini and Gammie.
Figures

Similar articles
-
Enabling Systemic Identification and Functionality Profiling for Cdc42 Homeostatic Modulators.bioRxiv [Preprint]. 2024 Jan 8:2024.01.05.574351. doi: 10.1101/2024.01.05.574351. bioRxiv. 2024. Update in: Commun Chem. 2024 Nov 19;7(1):271. doi: 10.1038/s42004-024-01352-7. PMID: 38260445 Free PMC article. Updated. Preprint.
-
The Smc5/6 complex counteracts R-loop formation at highly transcribed genes in cooperation with RNase H2.Elife. 2024 Oct 15;13:e96626. doi: 10.7554/eLife.96626. Elife. 2024. PMID: 39404251 Free PMC article.
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Interventions to reduce harm from continued tobacco use.Cochrane Database Syst Rev. 2016 Oct 13;10(10):CD005231. doi: 10.1002/14651858.CD005231.pub3. Cochrane Database Syst Rev. 2016. PMID: 27734465 Free PMC article. Review.
Cited by
-
miR-1266 Contributes to Pancreatic Cancer Progression and Chemoresistance by the STAT3 and NF-κB Signaling Pathways.Mol Ther Nucleic Acids. 2018 Jun 1;11:142-158. doi: 10.1016/j.omtn.2018.01.004. Epub 2018 Jan 31. Mol Ther Nucleic Acids. 2018. PMID: 29858050 Free PMC article.
-
The AKT inhibitor, MK-2206, attenuates ABCG2-mediated drug resistance in lung and colon cancer cells.Front Pharmacol. 2023 Jul 13;14:1235285. doi: 10.3389/fphar.2023.1235285. eCollection 2023. Front Pharmacol. 2023. PMID: 37521473 Free PMC article.
-
Comparative chemical genomics reveal that the spiroindolone antimalarial KAE609 (Cipargamin) is a P-type ATPase inhibitor.Sci Rep. 2016 Jun 13;6:27806. doi: 10.1038/srep27806. Sci Rep. 2016. PMID: 27291296 Free PMC article.
-
Finding the needle in a haystack: Mapping antifungal drug resistance in fungal pathogen by genomic approaches.PLoS Pathog. 2019 Jan 31;15(1):e1007478. doi: 10.1371/journal.ppat.1007478. eCollection 2019 Jan. PLoS Pathog. 2019. PMID: 30703166 Free PMC article. Review. No abstract available.
-
Downregulation of miR-374b-5p promotes chemotherapeutic resistance in pancreatic cancer by upregulating multiple anti-apoptotic proteins.Int J Oncol. 2018 May;52(5):1491-1503. doi: 10.3892/ijo.2018.4315. Epub 2018 Mar 14. Int J Oncol. 2018. PMID: 29568910 Free PMC article.
References
-
- Aebi S., Fink D., Gordon R., Kim H. K., Zheng H., et al. , 1997. Resistance to cytotoxic drugs in DNA mismatch repair-deficient cells. Clin. Cancer Res. 3: 1763–1767. - PubMed
-
- Agarwal R., Kaye S. B., 2003. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat. Rev. Cancer 3: 502–516. - PubMed
-
- Aouida M., Page N., Leduc A., Peter M., Ramotar D., 2004. A genome-wide screen in Saccharomyces cerevisiae reveals altered transport as a mechanism of resistance to the anticancer drug bleomycin. Cancer Res. 64: 1102–1109. - PubMed
-
- Arceci R. J., Stieglitz K., Bierer B. E., 1992. Immunosuppressants FK506 and rapamycin function as reversal agents of the multidrug resistance phenotype. Blood 80: 1528–1536. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
