lobChIP: from cells to sequencing ready ChIP libraries in a single day
- PMID: 26195988
- PMCID: PMC4507313
- DOI: 10.1186/s13072-015-0017-5
lobChIP: from cells to sequencing ready ChIP libraries in a single day
Abstract
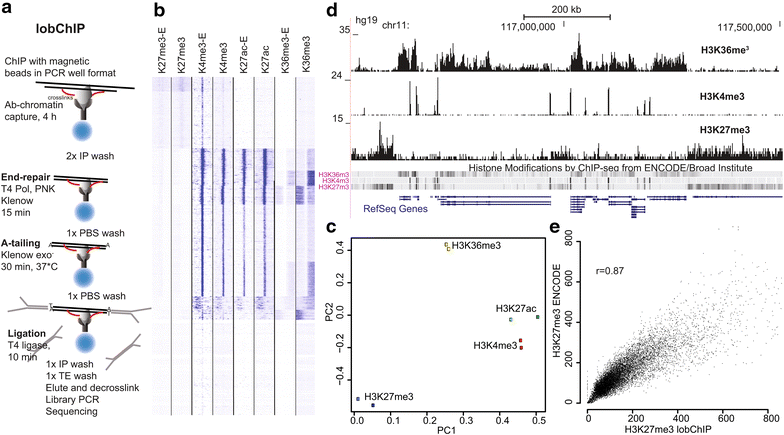
Background: ChIP-seq is the method of choice for genome-wide studies of protein-DNA interactions. We describe a new method for ChIP-seq sample preparation, termed lobChIP, where the library reactions are performed on cross-linked ChIP fragments captured on beads.
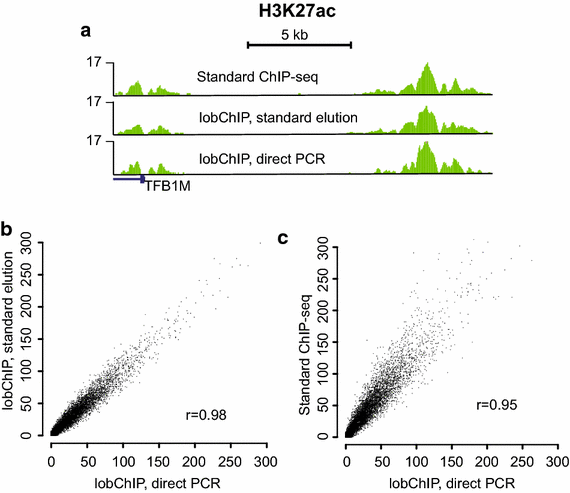
Results: The lobChIP method was found both to reduce time and cost and to simplify the processing of many samples in parallel. lobChIP has an early incorporation of barcoded sequencing adaptors that minimizes the risk of sample cross-contamination and can lead to reduced amount of adaptor dimers in the sequencing libraries, while allowing for direct decross-linking and amplification of the sample.
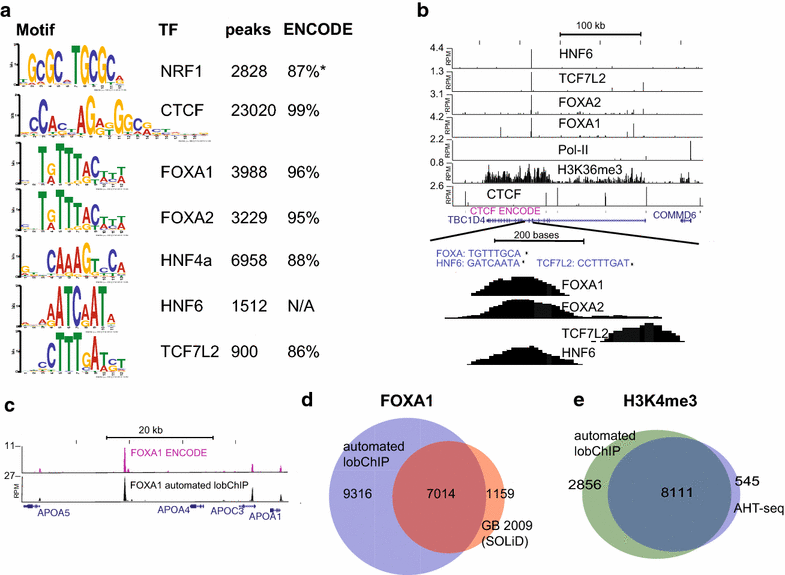
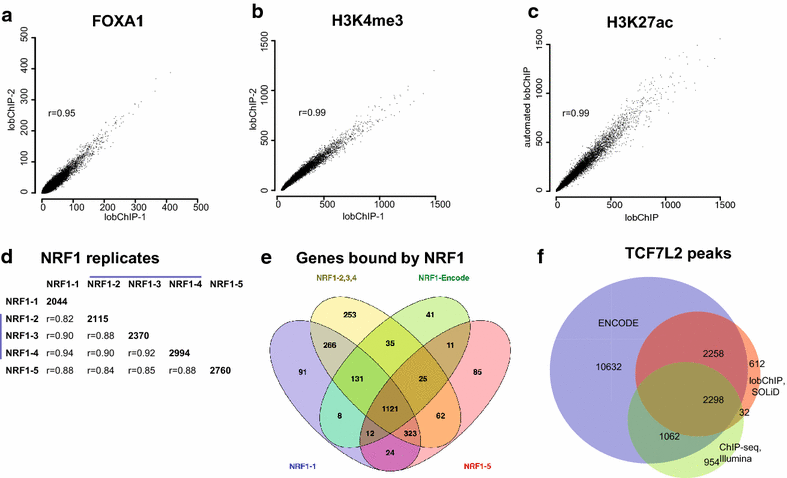
Conclusions: With results for histone modifications and transcription factors, we show that lobChIP performs equal to or better than standard protocols and that it makes it possible to go from cells to sequencing ready libraries within a single day.
Keywords: ChIP-seq; Chromatin immunoprecipitation; Illumina; Library preparation; NGS.
Figures
Similar articles
-
Preparation of Low-Input and Ligation-Free ChIP-seq Libraries Using Template-Switching Technology.Curr Protoc Mol Biol. 2016 Oct 10;116:7.28.1-7.28.26. doi: 10.1002/cpmb.24. Curr Protoc Mol Biol. 2016. PMID: 27723085
-
Chromatin analyses of Zymoseptoria tritici: Methods for chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq).Fungal Genet Biol. 2015 Jun;79:63-70. doi: 10.1016/j.fgb.2015.03.006. Epub 2015 Apr 7. Fungal Genet Biol. 2015. PMID: 25857259 Free PMC article.
-
ChIP-Seq using high-throughput DNA sequencing for genome-wide identification of transcription factor binding sites.Methods Enzymol. 2010;470:77-104. doi: 10.1016/S0076-6879(10)70004-5. Epub 2010 Mar 1. Methods Enzymol. 2010. PMID: 20946807
-
Multiplexed chromatin immunoprecipitation sequencing for quantitative study of histone modifications and chromatin factors.Nat Protoc. 2025 Mar;20(3):779-809. doi: 10.1038/s41596-024-01058-z. Epub 2024 Oct 3. Nat Protoc. 2025. PMID: 39363107 Review.
-
Revolution of nephrology research by deep sequencing: ChIP-seq and RNA-seq.Kidney Int. 2014 Jan;85(1):31-8. doi: 10.1038/ki.2013.321. Epub 2013 Aug 28. Kidney Int. 2014. PMID: 23986147 Review.
Cited by
-
High-throughput ChIPmentation: freely scalable, single day ChIPseq data generation from very low cell-numbers.BMC Genomics. 2019 Jan 18;20(1):59. doi: 10.1186/s12864-018-5299-0. BMC Genomics. 2019. PMID: 30658577 Free PMC article.
-
Changes in H3K27ac following lipopolysaccharide stimulation of nasopharyngeal epithelial cells.BMC Genomics. 2018 Dec 27;19(1):969. doi: 10.1186/s12864-018-5295-4. BMC Genomics. 2018. PMID: 30587130 Free PMC article.
-
PAtCh-Cap: input strategy for improving analysis of ChIP-exo data sets and beyond.Nucleic Acids Res. 2016 Dec 1;44(21):e159. doi: 10.1093/nar/gkw741. Epub 2016 Aug 22. Nucleic Acids Res. 2016. PMID: 27550178 Free PMC article.
-
Genome-wide histone modification profiling of inner cell mass and trophectoderm of bovine blastocysts by RAT-ChIP.PLoS One. 2019 Nov 25;14(11):e0225801. doi: 10.1371/journal.pone.0225801. eCollection 2019. PLoS One. 2019. PMID: 31765427 Free PMC article.
-
Genome-wide epigenomic profiling for biomarker discovery.Clin Epigenetics. 2016 Nov 21;8:122. doi: 10.1186/s13148-016-0284-4. eCollection 2016. Clin Epigenetics. 2016. PMID: 27895806 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

