Actin age orchestrates myosin-5 and myosin-6 run lengths
- PMID: 26190073
- PMCID: PMC4556227
- DOI: 10.1016/j.cub.2015.06.033
Actin age orchestrates myosin-5 and myosin-6 run lengths
Abstract
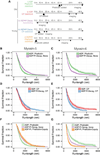
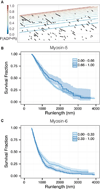
Unlike a static and immobile skeleton, the actin cytoskeleton is a highly dynamic network of filamentous actin (F-actin) polymers that continuously turn over. In addition to generating mechanical forces and sensing mechanical deformation, dynamic F-actin networks serve as cellular tracks for myosin motor traffic. However, much of our mechanistic understanding of processive myosins comes from in vitro studies in which motility was studied on pre-assembled and artificially stabilized, static F-actin tracks. In this work, we examine the role of actin dynamics in single-molecule myosin motility using assembling F-actin and two highly processive motors, myosin-5 and myosin-6. These two myosins have distinct functions in the cell and travel in opposite directions along actin filaments [1-3]. Myosin-5 walks toward the barbed ends of F-actin, traveling to sites of actin polymerization at the cell periphery [4]. Myosin-6 walks toward the pointed end of F-actin [5], traveling toward the cell center along older segments of the actin filament. We find that myosin-5 takes 1.3- to 1.5-fold longer runs on ADP•Pi (young) F-actin, whereas myosin-6 takes 1.7- to 3.6-fold longer runs along ADP (old) F-actin. These results suggest that conformational differences between ADP•Pi and ADP F-actin tailor these myosins to walk farther toward their preferred actin filament end. Taken together, these experiments define a new mechanism by which myosin traffic may sort to different F-actin networks depending on filament age.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
The stepping pattern of myosin X is adapted for processive motility on bundled actin.Biophys J. 2010 Sep 22;99(6):1818-26. doi: 10.1016/j.bpj.2010.06.066. Biophys J. 2010. PMID: 20858426 Free PMC article.
-
Myosin-Specific Adaptations of In vitro Fluorescence Microscopy-Based Motility Assays.J Vis Exp. 2021 Feb 4;(168). doi: 10.3791/62180. J Vis Exp. 2021. PMID: 33616114
-
Actin dynamics is essential for myosin-based transport of membrane organelles.Curr Biol. 2008 Oct 28;18(20):1581-6. doi: 10.1016/j.cub.2008.08.070. Curr Biol. 2008. PMID: 18951026 Free PMC article.
-
Cargo recognition and cargo-mediated regulation of unconventional myosins.Acc Chem Res. 2014 Oct 21;47(10):3061-70. doi: 10.1021/ar500216z. Epub 2014 Sep 17. Acc Chem Res. 2014. PMID: 25230296 Review.
-
How Actin Tracks Affect Myosin Motors.Adv Exp Med Biol. 2020;1239:183-197. doi: 10.1007/978-3-030-38062-5_9. Adv Exp Med Biol. 2020. PMID: 32451860 Review.
Cited by
-
Dynamics of Vesicles Driven Into Closed Constrictions by Molecular Motors.Bull Math Biol. 2020 Oct 23;82(11):141. doi: 10.1007/s11538-020-00820-0. Bull Math Biol. 2020. PMID: 33095297 Free PMC article.
-
Coordinated recruitment of Spir actin nucleators and myosin V motors to Rab11 vesicle membranes.Elife. 2016 Sep 13;5:e17523. doi: 10.7554/eLife.17523. Elife. 2016. PMID: 27623148 Free PMC article.
-
Motor domain phosphorylation increases nucleotide exchange and turns MYO6 into a faster and stronger motor.Nat Commun. 2024 Aug 7;15(1):6716. doi: 10.1038/s41467-024-49898-3. Nat Commun. 2024. PMID: 39112473 Free PMC article.
-
Causes, costs and consequences of kinesin motors communicating through the microtubule lattice.J Cell Sci. 2023 Mar 1;136(5):jcs260735. doi: 10.1242/jcs.260735. Epub 2023 Mar 3. J Cell Sci. 2023. PMID: 36866642 Free PMC article.
-
Cryo-EM structures reveal specialization at the myosin VI-actin interface and a mechanism of force sensitivity.Elife. 2017 Dec 4;6:e31125. doi: 10.7554/eLife.31125. Elife. 2017. PMID: 29199952 Free PMC article.
References
-
- Buss F, Spudich G, Kendrick-Jones J. Myosin VI: cellular functions and motor properties. Annu Rev Cell Dev Biol. 2004;20:649–676. - PubMed
-
- Sellers JR, Goodson HV. Motor proteins 2: myosin. Protein Profile. 1995;2:1323–1423. - PubMed
-
- Reck-Peterson SL, Provance DW, Mooseker MS, Mercer JA. Class V myosins. Biochim Biophys Acta. 2000;1496:36–51. - PubMed
-
- Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

