The Role of Protein-Ligand Contacts in Allosteric Regulation of the Escherichia coli Catabolite Activator Protein
- PMID: 26187469
- PMCID: PMC4571973
- DOI: 10.1074/jbc.M115.669267
The Role of Protein-Ligand Contacts in Allosteric Regulation of the Escherichia coli Catabolite Activator Protein
Abstract
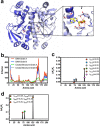
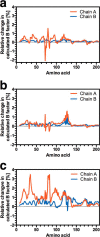
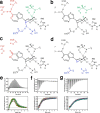
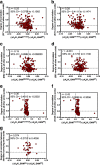
Allostery is a fundamental process by which ligand binding to a protein alters its activity at a distant site. Both experimental and theoretical evidence demonstrate that allostery can be communicated through altered slow relaxation protein dynamics without conformational change. The catabolite activator protein (CAP) of Escherichia coli is an exemplar for the analysis of such entropically driven allostery. Negative allostery in CAP occurs between identical cAMP binding sites. Changes to the cAMP-binding pocket can therefore impact the allosteric properties of CAP. Here we demonstrate, through a combination of coarse-grained modeling, isothermal calorimetry, and structural analysis, that decreasing the affinity of CAP for cAMP enhances negative cooperativity through an entropic penalty for ligand binding. The use of variant cAMP ligands indicates the data are not explained by structural heterogeneity between protein mutants. We observe computationally that altered interaction strength between CAP and cAMP variously modifies the change in allosteric cooperativity due to second site CAP mutations. As the degree of correlated motion between the cAMP-contacting site and a second site on CAP increases, there is a tendency for computed double mutations at these sites to drive CAP toward noncooperativity. Naturally occurring pairs of covarying residues in CAP do not display this tendency, suggesting a selection pressure to fine tune allostery on changes to the CAP ligand-binding pocket without a drive to a noncooperative state. In general, we hypothesize an evolutionary selection pressure to retain slow relaxation dynamics-induced allostery in proteins in which evolution of the ligand-binding site is occurring.
Keywords: allosteric regulation; biophysics; calorimetry; cyclic AMP (cAMP); nucleoside/nucleotide analogue; protein dynamic.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Modulation of global low-frequency motions underlies allosteric regulation: demonstration in CRP/FNR family transcription factors.PLoS Biol. 2013 Sep;11(9):e1001651. doi: 10.1371/journal.pbio.1001651. Epub 2013 Sep 10. PLoS Biol. 2013. PMID: 24058293 Free PMC article.
-
A computational investigation of allostery in the catabolite activator protein.J Am Chem Soc. 2007 Dec 19;129(50):15668-76. doi: 10.1021/ja076046a. Epub 2007 Nov 28. J Am Chem Soc. 2007. PMID: 18041838
-
Dynamically driven protein allostery.Nat Struct Mol Biol. 2006 Sep;13(9):831-8. doi: 10.1038/nsmb1132. Epub 2006 Aug 13. Nat Struct Mol Biol. 2006. PMID: 16906160 Free PMC article.
-
Syn, anti, and finally both conformations of cyclic AMP are involved in the CRP-dependent transcription initiation mechanism in E. coli lac operon.Cell Biochem Funct. 2008 Jun;26(4):399-405. doi: 10.1002/cbf.1462. Cell Biochem Funct. 2008. PMID: 18338329 Review.
-
Role of allosteric changes in cyclic AMP receptor protein function.Subcell Biochem. 1995;24:303-21. doi: 10.1007/978-1-4899-1727-0_10. Subcell Biochem. 1995. PMID: 7900180 Review. No abstract available.
Cited by
-
Small Molecule Targeting of Protein-Protein Interactions through Allosteric Modulation of Dynamics.Molecules. 2015 Sep 10;20(9):16435-45. doi: 10.3390/molecules200916435. Molecules. 2015. PMID: 26378508 Free PMC article. Review.
-
Allosteric rescue of catalytically impaired ATP phosphoribosyltransferase variants links protein dynamics to active-site electrostatic preorganisation.Nat Commun. 2022 Dec 9;13(1):7607. doi: 10.1038/s41467-022-34960-9. Nat Commun. 2022. PMID: 36494361 Free PMC article.
-
Dynamic Transmission of Protein Allostery without Structural Change: Spatial Pathways or Global Modes?Biophys J. 2015 Sep 15;109(6):1240-50. doi: 10.1016/j.bpj.2015.08.009. Epub 2015 Aug 31. Biophys J. 2015. PMID: 26338443 Free PMC article.
-
Energetic redistribution in allostery to execute protein function.Proc Natl Acad Sci U S A. 2017 Jul 18;114(29):7480-7482. doi: 10.1073/pnas.1709071114. Epub 2017 Jul 10. Proc Natl Acad Sci U S A. 2017. PMID: 28696318 Free PMC article. No abstract available.
-
Nucleotide Second Messenger-Based Signaling in Extreme Acidophiles of the Acidithiobacillus Species Complex: Partition Between the Core and Variable Gene Complements.Front Microbiol. 2019 Mar 7;10:381. doi: 10.3389/fmicb.2019.00381. eCollection 2019. Front Microbiol. 2019. PMID: 30899248 Free PMC article.
References
-
- Changeux J. P., Edelstein S. J. (2005) Allosteric mechanisms of signal transduction. Science 308, 1424–1428 - PubMed
-
- Koshland D. E. Jr., Némethy G., Filmer D. (1966) Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5, 365–385 - PubMed
-
- Monod J., Wyman J., Changeux J. P. (1965) On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 12, 88–118 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

