The UBR-box and its relationship to binuclear RING-like treble clef zinc fingers
- PMID: 26185100
- PMCID: PMC4506424
- DOI: 10.1186/s13062-015-0066-5
The UBR-box and its relationship to binuclear RING-like treble clef zinc fingers
Abstract
Background: The N-end rule pathway is a part of the ubiquitin-dependent proteolytic system wherein N-recognin proteins recognize the amino terminal degradation signals (N-degrons) of the substrate. The type 1 N-degron recognizing UBR-box domain of the eukaryotic Arg/N-end rule pathway is known to possess a novel three-zinc-stabilized heart-shaped fold.
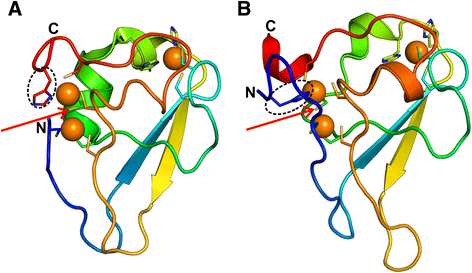
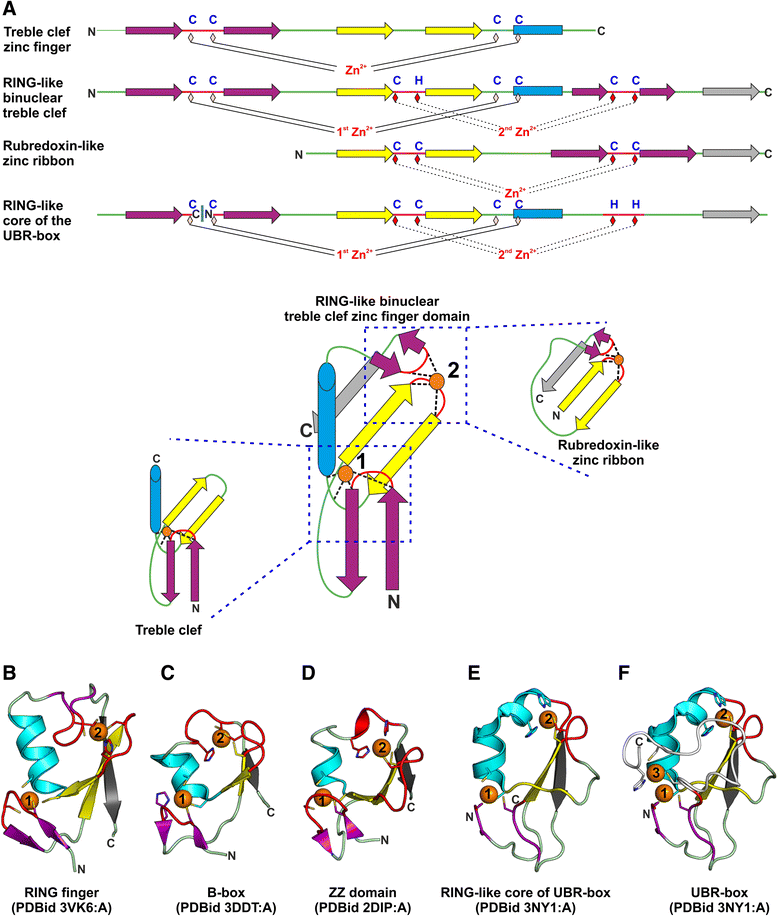
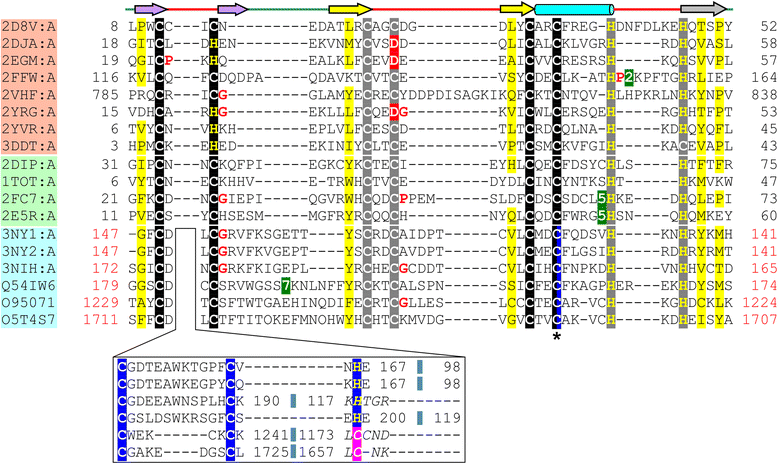
Results: Using sequence and structure analysis we argue that the UBR-box fold emerged from a binuclear RING-like treble clef zinc finger. The RING-like core is preserved in the UBR-box and the metal-chelating motifs display significant sequence and structural similarity to B-box and ZZ domains. UBR-box domains retrieved in our analysis co-occur with a variety of other protein domains, suggestive of its involvement in diverse biological roles. The UBR-box is a unique family of RING-like treble clefs as it displays a distinct circular permutation at the zinc-knuckle of the first zinc-binding site unlike other documented permutations of the RING-like domains which occur at the second zinc-binding site. The circular permutation of the RING-like treble clef scaffold has possibly aided the gain of a novel and relatively deep cleft suited for binding N-degrons. The N- and C-terminal extensions to the circularly permuted RING-like region bind a third zinc ion, which likely provides additional stability to the domain by keeping the two halves of the permuted zinc-knuckle together.
Conclusions: Structural modifications and extensions to the RING-like core have resulted in a novel UBR-box fold, which can recognize and target the type 1 N-degron containing proteins for ubiquitin-mediated proteolysis. The UBR-box appears to have emerged during the expansion of ubiquitin system pathway-related functions in eukaryotes, but is also likely to have other non-N-recognin functions as well.
Figures
Similar articles
-
A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons.Mol Cell Biol. 2005 Aug;25(16):7120-36. doi: 10.1128/MCB.25.16.7120-7136.2005. Mol Cell Biol. 2005. PMID: 16055722 Free PMC article.
-
Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system.Mol Biosyst. 2011 Jul;7(7):2261-77. doi: 10.1039/c1mb05061c. Epub 2011 May 6. Mol Biosyst. 2011. PMID: 21547297 Free PMC article.
-
The substrate recognition domains of the N-end rule pathway.J Biol Chem. 2009 Jan 16;284(3):1884-95. doi: 10.1074/jbc.M803641200. Epub 2008 Nov 13. J Biol Chem. 2009. PMID: 19008229 Free PMC article.
-
Signaling Pathways Regulated by UBR Box-Containing E3 Ligases.Int J Mol Sci. 2021 Aug 3;22(15):8323. doi: 10.3390/ijms22158323. Int J Mol Sci. 2021. PMID: 34361089 Free PMC article. Review.
-
How the ends signal the end: Regulation by E3 ubiquitin ligases recognizing protein termini.Mol Cell. 2022 Apr 21;82(8):1424-1438. doi: 10.1016/j.molcel.2022.02.004. Epub 2022 Mar 4. Mol Cell. 2022. PMID: 35247307 Free PMC article. Review.
Cited by
-
Evolutionary convergence and divergence in archaeal chromosomal proteins and Chromo-like domains from bacteria and eukaryotes.Sci Rep. 2018 Apr 18;8(1):6196. doi: 10.1038/s41598-018-24467-z. Sci Rep. 2018. PMID: 29670199 Free PMC article.
-
Insights into the recognition mechanism in the UBR box of UBR4 for its specific substrates.Commun Biol. 2023 Nov 29;6(1):1214. doi: 10.1038/s42003-023-05602-7. Commun Biol. 2023. PMID: 38030679 Free PMC article.
-
A novel RING finger in the C-terminal domain of the coatomer protein α-COP.Biol Direct. 2015 Dec 14;10:70. doi: 10.1186/s13062-015-0099-9. Biol Direct. 2015. PMID: 26666296 Free PMC article.
-
Classification of the treble clef zinc finger: noteworthy lessons for structure and function evolution.Sci Rep. 2016 Aug 26;6:32070. doi: 10.1038/srep32070. Sci Rep. 2016. PMID: 27562564 Free PMC article.
-
BIG Regulates Dynamic Adjustment of Circadian Period in Arabidopsis thaliana.Plant Physiol. 2018 Sep;178(1):358-371. doi: 10.1104/pp.18.00571. Epub 2018 Jul 11. Plant Physiol. 2018. PMID: 29997180 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

