Therapeutic doses of irradiation activate viral transcription and induce apoptosis in HIV-1 infected cells
- PMID: 26184775
- PMCID: PMC4619158
- DOI: 10.1016/j.virol.2015.06.021
Therapeutic doses of irradiation activate viral transcription and induce apoptosis in HIV-1 infected cells
Abstract
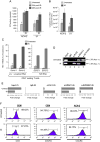
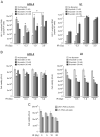
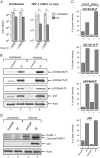
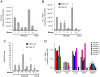
The highly active antiretroviral therapy reduces HIV-1 RNA in plasma to undetectable levels. However, the virus continues to persist in the long-lived resting CD4(+) T cells, macrophages and astrocytes which form a viral reservoir in infected individuals. Reactivation of viral transcription is critical since the host immune response in combination with antiretroviral therapy may eradicate the virus. Using the chronically HIV-1 infected T lymphoblastoid and monocytic cell lines, primary quiescent CD4(+) T cells and humanized mice infected with dual-tropic HIV-1 89.6, we examined the effect of various X-ray irradiation (IR) doses (used for HIV-related lymphoma treatment and lower doses) on HIV-1 transcription and viability of infected cells. Treatment of both T cells and monocytes with IR, a well-defined stress signal, led to increase of HIV-1 transcription, as evidenced by the presence of RNA polymerase II and reduction of HDAC1 and methyl transferase SUV39H1 on the HIV-1 promoter. This correlated with the increased GFP signal and elevated level of intracellular HIV-1 RNA in the IR-treated quiescent CD4(+) T cells infected with GFP-encoding HIV-1. Exposition of latently HIV-1infected monocytes treated with PKC agonist bryostatin 1 to IR enhanced transcription activation effect of this latency-reversing agent. Increased HIV-1 replication after IR correlated with higher cell death: the level of phosphorylated Ser46 in p53, responsible for apoptosis induction, was markedly higher in the HIV-1 infected cells following IR treatment. Exposure of HIV-1 infected humanized mice with undetectable viral RNA level to IR resulted in a significant increase of HIV-1 RNA in plasma, lung and brain tissues. Collectively, these data point to the use of low to moderate dose of IR alone or in combination with HIV-1 transcription activators as a potential application for the "Shock and Kill" strategy for latently HIV-1 infected cells.
Keywords: Apoptosis; CD4+ T cells; HIV-1 latency; HIV-1 reactivation; Humanized mice; Irradiation; Monocytes; Transcription; X-ray.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Potential of Radiation-Induced Cellular Stress for Reactivation of Latent HIV-1 and Killing of Infected Cells.AIDS Res Hum Retroviruses. 2016 Feb;32(2):120-4. doi: 10.1089/AID.2016.0006. AIDS Res Hum Retroviruses. 2016. PMID: 26765533 Free PMC article.
-
The Pathway To Establishing HIV Latency Is Critical to How Latency Is Maintained and Reversed.J Virol. 2018 Jun 13;92(13):e02225-17. doi: 10.1128/JVI.02225-17. Print 2018 Jul 1. J Virol. 2018. PMID: 29643247 Free PMC article.
-
In vivo analysis of the effect of panobinostat on cell-associated HIV RNA and DNA levels and latent HIV infection.Retrovirology. 2016 May 21;13(1):36. doi: 10.1186/s12977-016-0268-7. Retrovirology. 2016. PMID: 27206407 Free PMC article.
-
Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy.Annu Rev Immunol. 2000;18:665-708. doi: 10.1146/annurev.immunol.18.1.665. Annu Rev Immunol. 2000. PMID: 10837072 Review.
-
HIV-1 Transcription Inhibitors Increase the Synthesis of Viral Non-Coding RNA that Contribute to Latency.Curr Pharm Des. 2017;23(28):4133-4144. doi: 10.2174/1381612823666170622101319. Curr Pharm Des. 2017. PMID: 28641535 Review.
Cited by
-
HIV Preintegration Transcription and Host Antagonism.Curr HIV Res. 2023;21(3):160-171. doi: 10.2174/1570162X21666230621122637. Curr HIV Res. 2023. PMID: 37345240 Free PMC article.
-
Low-Level Ionizing Radiation Induces Selective Killing of HIV-1-Infected Cells with Reversal of Cytokine Induction Using mTOR Inhibitors.Viruses. 2020 Aug 13;12(8):885. doi: 10.3390/v12080885. Viruses. 2020. PMID: 32823598 Free PMC article.
-
Monitoring HIV DNA and cellular activation markers in HIV-infected humanized mice under cART.Virol J. 2018 Dec 17;15(1):191. doi: 10.1186/s12985-018-1101-9. Virol J. 2018. PMID: 30558630 Free PMC article.
-
Improvements and Limitations of Humanized Mouse Models for HIV Research: NIH/NIAID "Meet the Experts" 2015 Workshop Summary.AIDS Res Hum Retroviruses. 2016 Feb;32(2):109-19. doi: 10.1089/AID.2015.0258. AIDS Res Hum Retroviruses. 2016. PMID: 26670361 Free PMC article.
-
Inhibition of HIV-1 infection in humanized mice and metabolic stability of protein phosphatase-1-targeting small molecule 1E7-03.Oncotarget. 2017 Aug 7;8(44):76749-76769. doi: 10.18632/oncotarget.19999. eCollection 2017 Sep 29. Oncotarget. 2017. PMID: 29100346 Free PMC article.
References
-
- Aggarwal BB. Nuclear factor-kappaB: the enemy within. Cancer Cell. 2004;6:203–208. - PubMed
-
- Allers K, Hutter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, Schneider T. Evidence for the cure of HIV infection by CCR5Delta32/Delta32 stem cell transplantation. Blood. 2011;117:2791–2799. - PubMed
-
- Altschuler C, Resbeut M, Maraninchi D, Guillet JP, Blaise D, Stoppa AM, Carcassonne Y. Fractionated total body irradiation and allogeneic bone marrow transplantation for standard risk leukemia. Radiother Oncol. 1989;16:289–295. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

