Insights into the nuclear export of murine leukemia virus intron-containing RNA
- PMID: 26158194
- PMCID: PMC4615294
- DOI: 10.1080/15476286.2015.1065375
Insights into the nuclear export of murine leukemia virus intron-containing RNA
Abstract
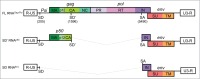
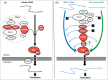
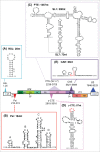
The retroviral genome consists of an intron-containing transcript that has essential cytoplasmic functions in the infected cell. This viral transcript can escape splicing, circumvent the nuclear checkpoint mechanisms and be transported to the cytoplasm by hijacking the host machinery. Once in the cytoplasm, viral unspliced RNA acts as mRNA to be translated and as genomic RNA to be packaged into nascent viruses. The murine leukemia virus (MLV) is among the first retroviruses discovered and is classified as simple Retroviridae due to its minimal encoding capacity. The oncogenic and transduction abilities of MLV are extensively studied, whereas surprisingly the crucial step of its nuclear export has remained unsolved until 2014. Recent work has revealed the recruitment by MLV of the cellular NXF1/Tap-dependent pathway for export. Unconventionally, MLV uses of Tap to export both spliced and unspliced viral RNAs. Unlike other retroviruses, MLV does not harbor a unique RNA signal for export. Indeed, multiple sequences throughout the MLV genome appear to promote export of the unspliced MLV RNA. We review here the current understanding of the export mechanism and highlight the determinants that influence MLV export. As the molecular mechanism of MLV export is elucidated, we will gain insight into the contribution of the export pathway to the cytoplasmic fate of the viral RNA.
Keywords: MLV; NXF1/Tap; RNA export; cis-acting RNA element; gammaretrovirus; intron retention; intron-containing RNA; nucleocytoplasmic trafficking; post-transcriptional gene regulation; virus replication.
Figures
Similar articles
-
NXF1 and CRM1 nuclear export pathways orchestrate nuclear export, translation and packaging of murine leukaemia retrovirus unspliced RNA.RNA Biol. 2020 Apr;17(4):528-538. doi: 10.1080/15476286.2020.1713539. Epub 2020 Jan 23. RNA Biol. 2020. PMID: 31918596 Free PMC article.
-
MLV requires Tap/NXF1-dependent pathway to export its unspliced RNA to the cytoplasm and to express both spliced and unspliced RNAs.Retrovirology. 2014 Mar 5;11:21. doi: 10.1186/1742-4690-11-21. Retrovirology. 2014. PMID: 24597485 Free PMC article.
-
Murine leukemia virus uses NXF1 for nuclear export of spliced and unspliced viral transcripts.J Virol. 2014 Apr;88(8):4069-82. doi: 10.1128/JVI.03584-13. Epub 2014 Jan 29. J Virol. 2014. PMID: 24478440 Free PMC article.
-
Retroviral RNA elements integrate components of post-transcriptional gene expression.Life Sci. 2001 Oct 26;69(23):2697-709. doi: 10.1016/s0024-3205(01)01360-1. Life Sci. 2001. PMID: 11720075 Review.
-
Biogenesis, assembly, and export of viral messenger ribonucleoproteins in the influenza A virus infected cell.RNA Biol. 2013 Aug;10(8):1274-82. doi: 10.4161/rna.25356. Epub 2013 Jun 17. RNA Biol. 2013. PMID: 23807439 Free PMC article. Review.
Cited by
-
Strength in Diversity: Nuclear Export of Viral RNAs.Viruses. 2020 Sep 11;12(9):1014. doi: 10.3390/v12091014. Viruses. 2020. PMID: 32932882 Free PMC article. Review.
-
Nuclear export of plant pararetrovirus mRNAs involves the TREX complex, two viral proteins and the highly structured 5' leader region.Nucleic Acids Res. 2021 Sep 7;49(15):8900-8922. doi: 10.1093/nar/gkab653. Nucleic Acids Res. 2021. PMID: 34370034 Free PMC article.
-
Recombinant origin and interspecies transmission of a HERV-K(HML-2)-related primate retrovirus with a novel RNA transport element.Elife. 2024 Jul 22;13:e80216. doi: 10.7554/eLife.80216. Elife. 2024. PMID: 39037763 Free PMC article.
-
NXF1 and CRM1 nuclear export pathways orchestrate nuclear export, translation and packaging of murine leukaemia retrovirus unspliced RNA.RNA Biol. 2020 Apr;17(4):528-538. doi: 10.1080/15476286.2020.1713539. Epub 2020 Jan 23. RNA Biol. 2020. PMID: 31918596 Free PMC article.
-
The piRNA Response to Retroviral Invasion of the Koala Genome.Cell. 2019 Oct 17;179(3):632-643.e12. doi: 10.1016/j.cell.2019.09.002. Epub 2019 Oct 10. Cell. 2019. PMID: 31607510 Free PMC article.
References
-
- Braoudaki M, Tzortzatou-Stathopoulou F. Tumorigenesis related to retroviral infections. J Infect Dev Ctries 2011; 5:751-8; PMID:22112727; http://dx.doi.org/10.3855/jidc.1773 - DOI - PubMed
-
- Wolff L, Koller R, Hu X, Anver MR. A Moloney murine leukemia virus-based retrovirus with 4070A long terminal repeat sequences induces a high incidence of myeloid as well as lymphoid neoplasms. J Virol 2003; 77:4965-71; PMID:12663802; http://dx.doi.org/10.1128/JVI.77.8.4965-4971.2003 - DOI - PMC - PubMed
-
- Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, Naiman DQ, Jenkins NA, Copeland NG. New genes involved in cancer identified by retroviral tagging. Nat Genet 2002; 32:166-74; PMID:12185365; http://dx.doi.org/10.1038/ng949 - DOI - PubMed
-
- Lund AH, Turner G, Trubetskoy A, Verhoeven E, Wientjens E, Hulsman D, Russell R, DePinho RA, Lenz J, Van Lohuizen M. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat Genet 2002; 32:160-5; PMID:12185367; http://dx.doi.org/10.1038/ng956 - DOI - PubMed
-
- Kim R, Trubetskoy A, Suzuki T, Jenkins NA, Copeland NG, Lenz J. Genome-based identification of cancer genes by proviral tagging in mouse retrovirus-induced T-cell lymphomas. J Virol 2003; 77:2056-62; PMID:12525640; http://dx.doi.org/10.1128/JVI.77.3.2056-2062.2003 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
