Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients
- PMID: 26151645
- PMCID: PMC4681870
- DOI: 10.1038/ismej.2015.99
Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients
Abstract
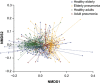
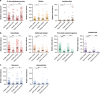
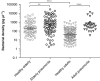
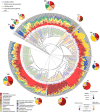
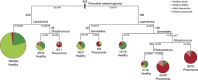
Bacterial pneumonia is a major cause of morbidity and mortality in elderly. We hypothesize that dysbiosis between regular residents of the upper respiratory tract (URT) microbiome, that is balance between commensals and potential pathogens, is involved in pathogen overgrowth and consequently disease. We compared oropharyngeal microbiota of elderly pneumonia patients (n=100) with healthy elderly (n=91) by 16S-rRNA-based sequencing and verified our findings in young adult pneumonia patients (n=27) and young healthy adults (n=187). Microbiota profiles differed significantly between elderly pneumonia patients and healthy elderly (PERMANOVA, P<0.0005). Highly similar differences were observed between microbiota profiles of young adult pneumonia patients and their healthy controls. Clustering resulted in 11 (sub)clusters including 95% (386/405) of samples. We observed three microbiota profiles strongly associated with pneumonia (P<0.05) and either dominated by lactobacilli (n=11), Rothia (n=51) or Streptococcus (pseudo)pneumoniae (n=42). In contrast, three other microbiota clusters (in total n=183) were correlated with health (P<0.05) and were all characterized by more diverse profiles containing higher abundances of especially Prevotella melaninogenica, Veillonella and Leptotrichia. For the remaining clusters (n=99), the association with health or disease was less clear. A decision tree model based on the relative abundance of five bacterial community members in URT microbiota showed high specificity of 95% and sensitivity of 84% (89% and 73%, respectively, after cross-validation) for differentiating pneumonia patients from healthy individuals. These results suggest that pneumonia in elderly and young adults is associated with dysbiosis of the URT microbiome with bacterial overgrowth of single species and absence of distinct anaerobic bacteria. Whether the observed microbiome changes are a cause or a consequence of the development of pneumonia or merely coincide with disease status remains a question for future research.
Conflict of interest statement
EAMS declares to have received unrestricted research support from Pfizer, grant support for vaccine studies from Pfizer and GlaxoSmithKline and fees paid to the institution for advisory boards or participation in independent data monitoring committees for Pfizer and GSK. RHV reported receiving grant support from GlaxoSmithKline and Wyeth/Pfizer for vaccine studies and consulting fees from GlaxoSmithKline. KT received grant support and consulting fees from Pfizer. DB received consulting fees from Pfizer. These grants and fees were not received for the research described in this paper. The remaining authors declare no conflict of interest.
Figures
Similar articles
-
Oropharyngeal microbiome profiling and its association with age and heart failure in the elderly population from the northernmost province of China.Microbiol Spectr. 2024 Oct 3;12(10):e0021624. doi: 10.1128/spectrum.00216-24. Epub 2024 Aug 20. Microbiol Spectr. 2024. PMID: 39162522 Free PMC article.
-
Upper Respiratory Dysbiosis with a Facultative-dominated Ecotype in Advanced Lung Disease and Dynamic Change after Lung Transplant.Ann Am Thorac Soc. 2019 Nov;16(11):1383-1391. doi: 10.1513/AnnalsATS.201904-299OC. Ann Am Thorac Soc. 2019. PMID: 31415219 Free PMC article.
-
Unique microbial landscape in the human oropharynx during different types of acute respiratory tract infections.Microbiome. 2023 Jul 24;11(1):157. doi: 10.1186/s40168-023-01597-9. Microbiome. 2023. PMID: 37482605 Free PMC article.
-
The role of the local microbial ecosystem in respiratory health and disease.Philos Trans R Soc Lond B Biol Sci. 2015 Aug 19;370(1675):20140294. doi: 10.1098/rstb.2014.0294. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26150660 Free PMC article. Review.
-
Respiratory microbiome in mechanically ventilated patients: a narrative review.Intensive Care Med. 2021 Mar;47(3):292-306. doi: 10.1007/s00134-020-06338-2. Epub 2021 Feb 9. Intensive Care Med. 2021. PMID: 33559707 Free PMC article. Review.
Cited by
-
Insights into the role of the respiratory tract microbiome in defense against bacterial pneumonia.Curr Opin Microbiol. 2024 Feb;77:102428. doi: 10.1016/j.mib.2024.102428. Epub 2024 Jan 25. Curr Opin Microbiol. 2024. PMID: 38277901 Free PMC article. Review.
-
Oropharyngeal microbiome profiling and its association with age and heart failure in the elderly population from the northernmost province of China.Microbiol Spectr. 2024 Oct 3;12(10):e0021624. doi: 10.1128/spectrum.00216-24. Epub 2024 Aug 20. Microbiol Spectr. 2024. PMID: 39162522 Free PMC article.
-
16S Metagenomic Comparison of Plasmodium falciparum-Infected and Noninfected Anopheles gambiae and Anopheles funestus Microbiota from Senegal.Am J Trop Med Hyg. 2018 Dec;99(6):1489-1498. doi: 10.4269/ajtmh.18-0263. Epub 2018 Oct 18. Am J Trop Med Hyg. 2018. PMID: 30350766 Free PMC article.
-
Microbiota in health and diseases.Signal Transduct Target Ther. 2022 Apr 23;7(1):135. doi: 10.1038/s41392-022-00974-4. Signal Transduct Target Ther. 2022. PMID: 35461318 Free PMC article. Review.
-
The respiratory tract microbiome, the pathogen load, and clinical interventions define severity of bacterial pneumonia.Cell Rep Med. 2023 Sep 19;4(9):101167. doi: 10.1016/j.xcrm.2023.101167. Epub 2023 Aug 25. Cell Rep Med. 2023. PMID: 37633274 Free PMC article.
References
-
- Breiman L. (1984) Classification and regression trees. Chapman & Hall/CRC: New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

