Potentiation of Growth Inhibitory Responses of the mTOR Inhibitor Everolimus by Dual mTORC1/2 Inhibitors in Cultured Breast Cancer Cell Lines
- PMID: 26148118
- PMCID: PMC4492962
- DOI: 10.1371/journal.pone.0131400
Potentiation of Growth Inhibitory Responses of the mTOR Inhibitor Everolimus by Dual mTORC1/2 Inhibitors in Cultured Breast Cancer Cell Lines
Abstract
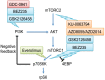
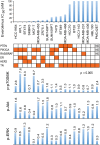
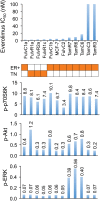
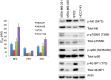
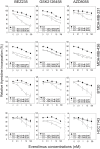
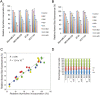
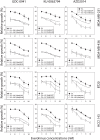
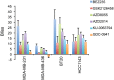
The mammalian target of rapamycin (mTOR), a vital component of signaling pathways involving PI3K/AKT, is an attractive therapeutic target in breast cancer. Everolimus, an allosteric mTOR inhibitor that inhibits the mTOR functional complex mTORC1, is approved for treatment of estrogen receptor positive (ER+) breast cancer. Other mTOR inhibitors show interesting differences in target specificities: BEZ235 and GSK2126458 are ATP competitive mTOR inhibitors targeting both PI3K and mTORC1/2; AZD8055, AZD2014 and KU-0063794 are ATP competitive mTOR inhibitors targeting both mTORC1 and mTORC2; and GDC-0941 is a pan-PI3K inhibitor. We have addressed the question of whether mTOR inhibitors may be more effective in combination than singly in inhibiting the proliferation of breast cancer cells. We selected a panel of 30 human breast cancer cell lines that included ER and PR positive, HER2 over-expressing, and "triple negative" variants, and determined whether signaling pathway utilization was related to drug-induced inhibition of proliferation. A significant correlation (p = 0.005) was found between everolimus IC50 values and p70S6K phosphorylation, but not with AKT or ERK phosphorylation, consistent with the mTOR pathway being a principal target. We then carried out combination studies with four everolimus resistant triple-negative breast cancer cell lines, and found an unexpectedly high degree of synergy between everolimus and the other inhibitors tested. The level of potentiation of everolimus inhibitory activity (measured by IC50 values) was found to be cell line-specific for all the kinase inhibitors tested. The results suggest that judicious combination of mTOR inhibitors with different modes of action could have beneficial effects in the treatment of breast cancer.
Conflict of interest statement
Figures

Similar articles
-
Impact of dual mTORC1/2 mTOR kinase inhibitor AZD8055 on acquired endocrine resistance in breast cancer in vitro.Breast Cancer Res. 2014 Jan 23;16(1):R12. doi: 10.1186/bcr3604. Breast Cancer Res. 2014. PMID: 24457069 Free PMC article.
-
Relationships between signaling pathway usage and sensitivity to a pathway inhibitor: examination of trametinib responses in cultured breast cancer lines.PLoS One. 2014 Aug 29;9(8):e105792. doi: 10.1371/journal.pone.0105792. eCollection 2014. PLoS One. 2014. PMID: 25170609 Free PMC article.
-
Dual inhibition of the PI3K/AKT/mTOR pathway suppresses the growth of leiomyosarcomas but leads to ERK activation through mTORC2: biological and clinical implications.Oncotarget. 2017 Jan 31;8(5):7878-7890. doi: 10.18632/oncotarget.13987. Oncotarget. 2017. PMID: 28002802 Free PMC article.
-
New inhibitors of the PI3K-Akt-mTOR pathway: insights into mTOR signaling from a new generation of Tor Kinase Domain Inhibitors (TORKinibs).Curr Top Microbiol Immunol. 2010;347:241-62. doi: 10.1007/82_2010_64. Curr Top Microbiol Immunol. 2010. PMID: 20549474 Review.
-
Evolving strategies for overcoming resistance to HER2-directed therapy: targeting the PI3K/Akt/mTOR pathway.Clin Breast Cancer. 2010 Nov;10 Suppl 3:S72-8. doi: 10.3816/CBC.2010.s.015. Clin Breast Cancer. 2010. PMID: 21115425 Review.
Cited by
-
Landscapes of cellular phenotypic diversity in breast cancer xenografts and their impact on drug response.Nat Commun. 2021 Mar 31;12(1):1998. doi: 10.1038/s41467-021-22303-z. Nat Commun. 2021. PMID: 33790302 Free PMC article.
-
The dual PI3K/mTOR inhibitor GSK2126458 is effective for treating solid renal tumours in Tsc2+/- mice through suppression of cell proliferation and induction of apoptosis.Oncotarget. 2017 Apr 19;8(35):58504-58512. doi: 10.18632/oncotarget.17215. eCollection 2017 Aug 29. Oncotarget. 2017. PMID: 28938574 Free PMC article.
-
Electroacupuncture protects against ischemic stroke by reducing autophagosome formation and inhibiting autophagy through the mTORC1-ULK1 complex-Beclin1 pathway.Int J Mol Med. 2016 Feb;37(2):309-18. doi: 10.3892/ijmm.2015.2425. Epub 2015 Dec 7. Int J Mol Med. 2016. PMID: 26647915 Free PMC article.
-
Molecular analysis of a male breast cancer patient with prolonged stable disease under mTOR/PI3K inhibitors BEZ235/everolimus.Cold Spring Harb Mol Case Stud. 2016 Mar;2(2):a000620. doi: 10.1101/mcs.a000620. Cold Spring Harb Mol Case Stud. 2016. PMID: 27148582 Free PMC article.
-
MNK1 inhibitor CGP57380 overcomes mTOR inhibitor-induced activation of eIF4E: the mechanism of synergic killing of human T-ALL cells.Acta Pharmacol Sin. 2018 Dec;39(12):1894-1901. doi: 10.1038/s41401-018-0161-0. Epub 2018 Oct 8. Acta Pharmacol Sin. 2018. PMID: 30297804 Free PMC article.
References
-
- Massarweh S, Romond E, Black EP, Van Meter E, Shelton B, Kadamyan-Melkumian V, et al. (2014) A phase II study of combined fulvestrant and everolimus in patients with metastatic estrogen receptor (ER)-positive breast cancer after aromatase inhibitor (AI) failure. Breast Cancer Res Treat 143: 325–332. 10.1007/s10549-013-2810-9 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

