Retroactivity in the Context of Modularly Structured Biomolecular Systems
- PMID: 26137457
- PMCID: PMC4470261
- DOI: 10.3389/fbioe.2015.00085
Retroactivity in the Context of Modularly Structured Biomolecular Systems
Abstract
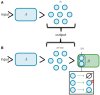
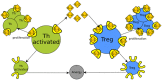
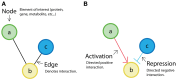
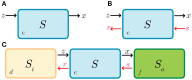
Synthetic biology has intensively promoted the technical implementation of modular strategies in the fabrication of biological devices. Modules are considered as networks of reactions. The behavior displayed by biomolecular systems results from the information processes carried out by the interconnection of the involved modules. However, in natural systems, module wiring is not a free-of-charge process; as a consequence of interconnection, a reactive phenomenon called retroactivity emerges. This phenomenon is characterized by signals that propagate from downstream modules (the modules that receive the incoming signals upon interconnection) to upstream ones (the modules that send the signals upon interconnection). Such retroactivity signals, depending of their strength, may change and sometimes even disrupt the behavior of modular biomolecular systems. Thus, analysis of retroactivity effects in natural biological and biosynthetic systems is crucial to achieve a deeper understanding of how this interconnection between functionally characterized modules takes place and how it impacts the overall behavior of the involved cell. By discussing the modules interconnection in natural and synthetic biomolecular systems, we propose that such systems should be considered as quasi-modular.
Keywords: modularity; regulatory biomolecular networks; retroactivity; signal transduction; synthetic biology; systems biology.
Figures


Similar articles
-
Modular composition of gene transcription networks.PLoS Comput Biol. 2014 Mar 13;10(3):e1003486. doi: 10.1371/journal.pcbi.1003486. eCollection 2014 Mar. PLoS Comput Biol. 2014. PMID: 24626132 Free PMC article.
-
A Practical Step-by-Step Guide for Quantifying Retroactivity in Gene Networks.Methods Mol Biol. 2021;2229:293-311. doi: 10.1007/978-1-0716-1032-9_14. Methods Mol Biol. 2021. PMID: 33405228
-
Modularity in signaling systems.Phys Biol. 2012 Aug;9(4):045008. doi: 10.1088/1478-3975/9/4/045008. Epub 2012 Aug 7. Phys Biol. 2012. PMID: 22871977
-
RTRACS: a modularized RNA-dependent RNA transcription system with high programmability.Acc Chem Res. 2011 Dec 20;44(12):1369-79. doi: 10.1021/ar200128b. Epub 2011 Oct 19. Acc Chem Res. 2011. PMID: 22011083 Review.
-
What Symbionts Teach us about Modularity.Front Bioeng Biotechnol. 2013 Nov 4;1:14. doi: 10.3389/fbioe.2013.00014. eCollection 2013. Front Bioeng Biotechnol. 2013. PMID: 25023877 Free PMC article. Review.
Cited by
-
RNAs competing for microRNAs mutually influence their fluctuations in a highly non-linear microRNA-dependent manner in single cells.Genome Biol. 2017 Feb 20;18(1):37. doi: 10.1186/s13059-017-1162-x. Genome Biol. 2017. PMID: 28219439 Free PMC article.
-
Automated design of synthetic microbial communities.Nat Commun. 2021 Jan 28;12(1):672. doi: 10.1038/s41467-020-20756-2. Nat Commun. 2021. PMID: 33510148 Free PMC article.
-
Context-aware synthetic biology by controller design: Engineering the mammalian cell.Cell Syst. 2021 Jun 16;12(6):561-592. doi: 10.1016/j.cels.2021.05.011. Cell Syst. 2021. PMID: 34139166 Free PMC article. Review.
-
From Microbial Communities to Distributed Computing Systems.Front Bioeng Biotechnol. 2020 Jul 22;8:834. doi: 10.3389/fbioe.2020.00834. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32793576 Free PMC article. Review.
-
Robust gene expression control in human cells with a novel universal TetR aptamer splicing module.Nucleic Acids Res. 2019 Nov 18;47(20):e132. doi: 10.1093/nar/gkz753. Nucleic Acids Res. 2019. PMID: 31504742 Free PMC article.
References
-
- Alon U. (2007). An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton, FL: CRC Press.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

