SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation
- PMID: 26134849
- PMCID: PMC4489885
- DOI: 10.1371/journal.pgen.1005367
SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation
Abstract
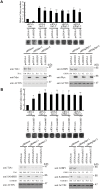
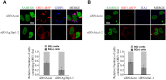
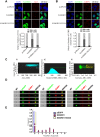
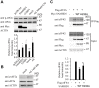
The SAM domain and HD domain containing protein 1 (SAMHD1) inhibits retroviruses, DNA viruses and long interspersed element 1 (LINE-1). Given that in dividing cells, SAMHD1 loses its antiviral function yet still potently restricts LINE-1, we propose that, instead of blocking viral DNA synthesis by virtue of its dNTP triphosphohydrolase activity, SAMHD1 may exploit a different mechanism to control LINE-1. Here, we report a new activity of SAMHD1 in promoting cellular stress granule assembly, which correlates with increased phosphorylation of eIF2α and diminished eIF4A/eIF4G interaction. This function of SAMHD1 enhances sequestration of LINE-1 RNP in stress granules and consequent blockade to LINE-1 retrotransposition. In support of this new mechanism of action, depletion of stress granule marker proteins G3BP1 or TIA1 abrogates stress granule formation and overcomes SAMHD1 inhibition of LINE-1. Together, these data reveal a new mechanism for SAMHD1 to control LINE-1 by activating cellular stress granule pathway.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
MxB inhibits long interspersed element type 1 retrotransposition.PLoS Genet. 2022 Feb 16;18(2):e1010034. doi: 10.1371/journal.pgen.1010034. eCollection 2022 Feb. PLoS Genet. 2022. PMID: 35171907 Free PMC article.
-
Hepatitis C virus (HCV) induces formation of stress granules whose proteins regulate HCV RNA replication and virus assembly and egress.J Virol. 2012 Oct;86(20):11043-56. doi: 10.1128/JVI.07101-11. Epub 2012 Aug 1. J Virol. 2012. PMID: 22855484 Free PMC article.
-
Seneca Valley Virus 3C Protease Inhibits Stress Granule Formation by Disrupting eIF4GI-G3BP1 Interaction.Front Immunol. 2020 Sep 29;11:577838. doi: 10.3389/fimmu.2020.577838. eCollection 2020. Front Immunol. 2020. PMID: 33133097 Free PMC article.
-
Cytoplasmic mRNP granules at a glance.J Cell Sci. 2011 Feb 1;124(Pt 3):293-7. doi: 10.1242/jcs.072140. J Cell Sci. 2011. PMID: 21242308 Free PMC article. Review. No abstract available.
-
[Research progress of the anti-HIV activity of SAMHD1].Bing Du Xue Bao. 2012 Jun;28(4):477-81. Bing Du Xue Bao. 2012. PMID: 22978176 Review. Chinese.
Cited by
-
Dual roles of SAMHD1 in tumor development and chemoresistance to anticancer drugs.Oncol Lett. 2021 Jun;21(6):451. doi: 10.3892/ol.2021.12712. Epub 2021 Apr 8. Oncol Lett. 2021. PMID: 33907561 Free PMC article. Review.
-
Single-Stranded Nucleic Acids Bind to the Tetramer Interface of SAMHD1 and Prevent Formation of the Catalytic Homotetramer.Biochemistry. 2016 Nov 8;55(44):6087-6099. doi: 10.1021/acs.biochem.6b00986. Epub 2016 Oct 27. Biochemistry. 2016. PMID: 27775344 Free PMC article.
-
Conservation and Innovation of APOBEC3A Restriction Functions during Primate Evolution.Mol Biol Evol. 2016 Aug;33(8):1889-901. doi: 10.1093/molbev/msw070. Epub 2016 Apr 6. Mol Biol Evol. 2016. PMID: 27189538 Free PMC article.
-
MxB inhibits long interspersed element type 1 retrotransposition.PLoS Genet. 2022 Feb 16;18(2):e1010034. doi: 10.1371/journal.pgen.1010034. eCollection 2022 Feb. PLoS Genet. 2022. PMID: 35171907 Free PMC article.
-
Ribonucleoprotein Granules: Between Stress and Transposable Elements.Biomolecules. 2023 Jun 23;13(7):1027. doi: 10.3390/biom13071027. Biomolecules. 2023. PMID: 37509063 Free PMC article. Review.
References
-
- Li N, Zhang W, Cao X (2000) Identification of human homologue of mouse IFN-gamma induced protein from human dendritic cells. Immunology letters 74: 221–224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

