Crystallographic structure of a small molecule SIRT1 activator-enzyme complex
- PMID: 26134520
- PMCID: PMC4506539
- DOI: 10.1038/ncomms8645
Crystallographic structure of a small molecule SIRT1 activator-enzyme complex
Abstract
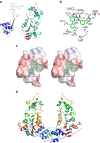
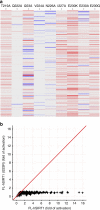
SIRT1, the founding member of the mammalian family of seven NAD(+)-dependent sirtuins, is composed of 747 amino acids forming a catalytic domain and extended N- and C-terminal regions. We report the design and characterization of an engineered human SIRT1 construct (mini-hSIRT1) containing the minimal structural elements required for lysine deacetylation and catalytic activation by small molecule sirtuin-activating compounds (STACs). Using this construct, we solved the crystal structure of a mini-hSIRT1-STAC complex, which revealed the STAC-binding site within the N-terminal domain of hSIRT1. Together with hydrogen-deuterium exchange mass spectrometry (HDX-MS) and site-directed mutagenesis using full-length hSIRT1, these data establish a specific STAC-binding site and identify key intermolecular interactions with hSIRT1. The determination of the interface governing the binding of STACs with human SIRT1 facilitates greater understanding of STAC activation of this enzyme, which holds significant promise as a therapeutic target for multiple human diseases.
Conflict of interest statement
H.D., A.W.C., T.V.R., T.C., H.Z., Y.J., S.M.S., B.P., B.S., C.A.B., J.S.D., R.C., B.S., C.O., P.Y.N., B.H.W., R.C., R.N., K.K., F.B., M.W., E.H., J.C.W., R.B.P., G.P.V. and J.L.E. are employees GlaxoSmithKline. Patent applications relating to the synthetic compounds have been filed.
Figures

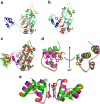

Similar articles
-
Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol.Genes Dev. 2015 Jun 15;29(12):1316-25. doi: 10.1101/gad.265462.115. Genes Dev. 2015. PMID: 26109052 Free PMC article.
-
The 2.5 Å crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition.J Med Chem. 2013 Feb 14;56(3):963-9. doi: 10.1021/jm301431y. Epub 2013 Jan 29. J Med Chem. 2013. PMID: 23311358
-
Multiscale landscape of molecular mechanism of SIRT1 activation by STACs.Phys Chem Chem Phys. 2020 Jan 2;22(2):826-837. doi: 10.1039/c9cp04931b. Phys Chem Chem Phys. 2020. PMID: 31840716
-
Sirtuin mechanism and inhibition: explored with N(ε)-acetyl-lysine analogs.Mol Biosyst. 2011 Jan;7(1):16-28. doi: 10.1039/c0mb00033g. Epub 2010 Sep 15. Mol Biosyst. 2011. PMID: 20842312 Review.
-
Small molecule SIRT1 activators for the treatment of aging and age-related diseases.Trends Pharmacol Sci. 2014 Mar;35(3):146-54. doi: 10.1016/j.tips.2013.12.004. Epub 2014 Jan 16. Trends Pharmacol Sci. 2014. PMID: 24439680 Free PMC article. Review.
Cited by
-
A molecular imaging biosensor detects in vivo protein folding and misfolding.J Mol Med (Berl). 2016 Jul;94(7):799-808. doi: 10.1007/s00109-016-1437-9. Epub 2016 Jun 8. J Mol Med (Berl). 2016. PMID: 27277823
-
Metformin Is a Direct SIRT1-Activating Compound: Computational Modeling and Experimental Validation.Front Endocrinol (Lausanne). 2018 Nov 6;9:657. doi: 10.3389/fendo.2018.00657. eCollection 2018. Front Endocrinol (Lausanne). 2018. PMID: 30459716 Free PMC article.
-
Exploration of stilbenoid trimers as potential inhibitors of sirtuin1 enzyme using a molecular docking and molecular dynamics simulation approach.RSC Adv. 2021 May 27;11(31):19323-19332. doi: 10.1039/d1ra02233d. eCollection 2021 May 24. RSC Adv. 2021. PMID: 35478645 Free PMC article.
-
Prediction and confirmation of a switch-like region within the N-terminal domain of hSIRT1.Biochem Biophys Rep. 2022 May 12;30:101275. doi: 10.1016/j.bbrep.2022.101275. eCollection 2022 Jul. Biochem Biophys Rep. 2022. PMID: 35592613 Free PMC article.
-
Myricanol rescues dexamethasone-induced muscle dysfunction via a sirtuin 1-dependent mechanism.J Cachexia Sarcopenia Muscle. 2019 Apr;10(2):429-444. doi: 10.1002/jcsm.12393. Epub 2019 Feb 21. J Cachexia Sarcopenia Muscle. 2019. PMID: 30793539 Free PMC article.
References
-
- Frye R. A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798 (2000). - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

