Characterization of ERM transactivation domain binding to the ACID/PTOV domain of the Mediator subunit MED25
- PMID: 26130716
- PMCID: PMC4538835
- DOI: 10.1093/nar/gkv650
Characterization of ERM transactivation domain binding to the ACID/PTOV domain of the Mediator subunit MED25
Abstract
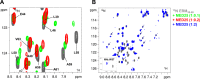
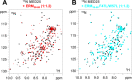
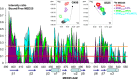
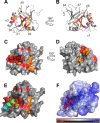
The N-terminal acidic transactivation domain (TAD) of ERM/ETV5 (ERM38-68), a PEA3 group member of Ets-related transcription factors, directly interacts with the ACID/PTOV domain of the Mediator complex subunit MED25. Molecular details of this interaction were investigated using nuclear magnetic resonance (NMR) spectroscopy. The TAD is disordered in solution but has a propensity to adopt local transient secondary structure. We show that it folds upon binding to MED25 and that the resulting ERM-MED25 complex displays characteristics of a fuzzy complex. Mutational analysis further reveals that two aromatic residues in the ERM TAD (F47 and W57) are involved in the binding to MED25 and participate in the ability of ERM TAD to activate transcription. Mutation of a key residue Q451 in the VP16 H1 binding pocket of MED25 affects the binding of ERM. Furthermore, competition experiments show that ERM and VP16 H1 share a common binding interface on MED25. NMR data confirms the occupancy of this binding pocket by ERM TAD. Based on these experimental data, a structural model of a functional interaction is proposed. This study provides mechanistic insights into the Mediator-transactivator interactions.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Structural Basis for the Interaction between p53 Transactivation Domain and the Mediator Subunit MED25.Molecules. 2018 Oct 22;23(10):2726. doi: 10.3390/molecules23102726. Molecules. 2018. PMID: 30360415 Free PMC article.
-
The Mediator complex subunit MED25 is targeted by the N-terminal transactivation domain of the PEA3 group members.Nucleic Acids Res. 2013 May;41(9):4847-59. doi: 10.1093/nar/gkt199. Epub 2013 Mar 26. Nucleic Acids Res. 2013. PMID: 23531547 Free PMC article.
-
NMR structure of the human Mediator MED25 ACID domain.J Struct Biol. 2011 Apr;174(1):245-51. doi: 10.1016/j.jsb.2010.10.011. Epub 2010 Oct 23. J Struct Biol. 2011. PMID: 20974256
-
The Mediator complex and the role of protein-protein interactions in the gene regulation machinery.Semin Cell Dev Biol. 2020 Mar;99:20-30. doi: 10.1016/j.semcdb.2018.08.006. Epub 2018 Oct 16. Semin Cell Dev Biol. 2020. PMID: 30278226 Review.
-
The Multitalented MEDIATOR25.Front Plant Sci. 2017 Jun 12;8:999. doi: 10.3389/fpls.2017.00999. eCollection 2017. Front Plant Sci. 2017. PMID: 28659948 Free PMC article. Review.
Cited by
-
Inhibition of CREB Binding and Function with a Dual-Targeting Ligand.Biochemistry. 2024 Jan 2;63(1):1-8. doi: 10.1021/acs.biochem.3c00469. Epub 2023 Dec 12. Biochemistry. 2024. PMID: 38086054 Free PMC article.
-
Molecular Dynamics of "Fuzzy" Transcriptional Activator-Coactivator Interactions.PLoS Comput Biol. 2016 May 13;12(5):e1004935. doi: 10.1371/journal.pcbi.1004935. eCollection 2016 May. PLoS Comput Biol. 2016. PMID: 27175900 Free PMC article.
-
Molecular switching in transcription through splicing and proline-isomerization regulates stress responses in plants.Nat Commun. 2024 Jan 18;15(1):592. doi: 10.1038/s41467-024-44859-2. Nat Commun. 2024. PMID: 38238333 Free PMC article.
-
ETV4 and AP1 Transcription Factors Form Multivalent Interactions with three Sites on the MED25 Activator-Interacting Domain.J Mol Biol. 2017 Oct 13;429(20):2975-2995. doi: 10.1016/j.jmb.2017.06.024. Epub 2017 Jul 17. J Mol Biol. 2017. PMID: 28728983 Free PMC article.
-
Structural Basis for the Interaction between p53 Transactivation Domain and the Mediator Subunit MED25.Molecules. 2018 Oct 22;23(10):2726. doi: 10.3390/molecules23102726. Molecules. 2018. PMID: 30360415 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

