Modeling cancer metabolism on a genome scale
- PMID: 26130389
- PMCID: PMC4501850
- DOI: 10.15252/msb.20145307
Modeling cancer metabolism on a genome scale
Abstract
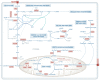
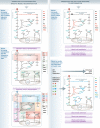
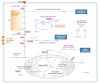
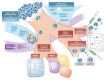
Cancer cells have fundamentally altered cellular metabolism that is associated with their tumorigenicity and malignancy. In addition to the widely studied Warburg effect, several new key metabolic alterations in cancer have been established over the last decade, leading to the recognition that altered tumor metabolism is one of the hallmarks of cancer. Deciphering the full scope and functional implications of the dysregulated metabolism in cancer requires both the advancement of a variety of omics measurements and the advancement of computational approaches for the analysis and contextualization of the accumulated data. Encouragingly, while the metabolic network is highly interconnected and complex, it is at the same time probably the best characterized cellular network. Following, this review discusses the challenges that genome-scale modeling of cancer metabolism has been facing. We survey several recent studies demonstrating the first strides that have been done, testifying to the value of this approach in portraying a network-level view of the cancer metabolism and in identifying novel drug targets and biomarkers. Finally, we outline a few new steps that may further advance this field.
Keywords: Cancer metabolism; Genome‐scale simulations; Metabolic modeling.
© 2015 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
Similar articles
-
Genome-scale modeling of human metabolism - a systems biology approach.Biotechnol J. 2013 Sep;8(9):985-96. doi: 10.1002/biot.201200275. Epub 2013 Apr 24. Biotechnol J. 2013. PMID: 23613448 Review.
-
Metabolic cancer biology: structural-based analysis of cancer as a metabolic disease, new sights and opportunities for disease treatment.Semin Cancer Biol. 2015 Feb;30:21-9. doi: 10.1016/j.semcancer.2014.01.007. Epub 2014 Feb 2. Semin Cancer Biol. 2015. PMID: 24495661 Review.
-
Big data in yeast systems biology.FEMS Yeast Res. 2019 Nov 1;19(7):foz070. doi: 10.1093/femsyr/foz070. FEMS Yeast Res. 2019. PMID: 31603503 Review.
-
Modeling cancer: integration of "omics" information in dynamic systems.J Bioinform Comput Biol. 2007 Aug;5(4):977-86. doi: 10.1142/s0219720007002990. J Bioinform Comput Biol. 2007. PMID: 17787066 Review.
-
Cancer Metabolism: A Modeling Perspective.Front Physiol. 2015 Dec 16;6:382. doi: 10.3389/fphys.2015.00382. eCollection 2015. Front Physiol. 2015. PMID: 26733270 Free PMC article. Review.
Cited by
-
Constraint-based modelling predicts metabolic signatures of low and high-grade serous ovarian cancer.NPJ Syst Biol Appl. 2024 Aug 24;10(1):96. doi: 10.1038/s41540-024-00418-5. NPJ Syst Biol Appl. 2024. PMID: 39181893 Free PMC article.
-
Modeling of Tumor Growth with Input from Patient-Specific Metabolomic Data.Ann Biomed Eng. 2022 Mar;50(3):314-329. doi: 10.1007/s10439-022-02904-5. Epub 2022 Jan 26. Ann Biomed Eng. 2022. PMID: 35083584 Free PMC article.
-
Common biochemical properties of metabolic genes recurrently dysregulated in tumors.Cancer Metab. 2020 May 8;8:5. doi: 10.1186/s40170-020-0211-1. eCollection 2020. Cancer Metab. 2020. PMID: 32411371 Free PMC article.
-
Personalized Prediction of Proliferation Rates and Metabolic Liabilities in Cancer Biopsies.Front Physiol. 2016 Dec 27;7:644. doi: 10.3389/fphys.2016.00644. eCollection 2016. Front Physiol. 2016. PMID: 28082911 Free PMC article.
-
Transcriptomics resources of human tissues and organs.Mol Syst Biol. 2016 Apr 4;12(4):862. doi: 10.15252/msb.20155865. Mol Syst Biol. 2016. PMID: 27044256 Free PMC article. Review.
References
-
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale A-L, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. - PMC - PubMed
-
- Alo PL, Visca P, Marci A, Mangoni A, Botti C, Di Tondo U. Expression of fatty acid synthase (FAS) as a predictor of recurrence in stage I breast carcinoma patients. Cancer. 1996;77:474–482. - PubMed
-
- Anesiadis N, Cluett WR, Mahadevan R. Dynamic metabolic engineering for increasing bioprocess productivity. Metab Eng. 2008;10:255–266. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

