The SEQUEST family tree
- PMID: 26122518
- PMCID: PMC4607603
- DOI: 10.1007/s13361-015-1201-3
The SEQUEST family tree
Abstract
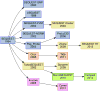
Since its introduction in 1994, SEQUEST has gained many important new capabilities, and a host of successor algorithms have built upon its successes. This Account and Perspective maps the evolution of this important tool and charts the relationships among contributions to the SEQUEST legacy. Many of the changes represented improvements in computing speed by clusters and graphics cards. Mass spectrometry innovations in mass accuracy and activation methods led to shifts in fragment modeling and scoring strategies. These changes, as well as the movement of laboratories and lab members, have led to great diversity among the members of the SEQUEST family. Graphical Abstract ᅟ.
Keywords: Bioinformatics; History; Intellectual property; Proteomics; SEQUEST.
Figures
Similar articles
-
Faster SEQUEST searching for peptide identification from tandem mass spectra.J Proteome Res. 2011 Sep 2;10(9):3871-9. doi: 10.1021/pr101196n. Epub 2011 Jul 29. J Proteome Res. 2011. PMID: 21761931 Free PMC article.
-
Pivotal role of computers and software in mass spectrometry - SEQUEST and 20 years of tandem MS database searching.J Am Soc Mass Spectrom. 2015 Nov;26(11):1804-13. doi: 10.1007/s13361-015-1220-0. Epub 2015 Aug 19. J Am Soc Mass Spectrom. 2015. PMID: 26286455 Free PMC article.
-
ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity.J Proteomics. 2015 Nov 3;129:16-24. doi: 10.1016/j.jprot.2015.07.001. Epub 2015 Jul 11. J Proteomics. 2015. PMID: 26171723 Free PMC article.
-
Protein identification using Sorcerer 2 and SEQUEST.Curr Protoc Bioinformatics. 2009 Dec;Chapter 13:Unit 13.3. doi: 10.1002/0471250953.bi1303s28. Curr Protoc Bioinformatics. 2009. PMID: 19957274 Review.
-
Free Open Source Software for Protein and Peptide Mass Spectrometry- based Science.Curr Protein Pept Sci. 2021;22(2):134-147. doi: 10.2174/1389203722666210118160946. Curr Protein Pept Sci. 2021. PMID: 33461461 Review.
Cited by
-
P-Mart: Interactive Analysis of Ion Abundance Global Proteomics Data.J Proteome Res. 2019 Mar 1;18(3):1426-1432. doi: 10.1021/acs.jproteome.8b00840. Epub 2019 Feb 6. J Proteome Res. 2019. PMID: 30667224 Free PMC article.
-
A multi-purpose, regenerable, proteome-scale, human phosphoserine resource for phosphoproteomics.Nat Methods. 2022 Nov;19(11):1371-1375. doi: 10.1038/s41592-022-01638-5. Epub 2022 Oct 24. Nat Methods. 2022. PMID: 36280721 Free PMC article.
-
BLM helicase protein negatively regulates stress granule formation through unwinding RNA G-quadruplex structures.Nucleic Acids Res. 2023 Sep 22;51(17):9369-9384. doi: 10.1093/nar/gkad613. Nucleic Acids Res. 2023. PMID: 37503837 Free PMC article.
-
Capillary pruning couples tissue perfusion and oxygenation with cardiomyocyte maturation in the postnatal mouse heart.Front Cell Dev Biol. 2023 Nov 7;11:1256127. doi: 10.3389/fcell.2023.1256127. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 38020883 Free PMC article.
-
A dynamic view of the proteomic landscape during differentiation of ReNcell VM cells, an immortalized human neural progenitor line.Sci Data. 2019 Feb 19;6:190016. doi: 10.1038/sdata.2019.16. Sci Data. 2019. PMID: 30778261 Free PMC article.
References
-
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. - PubMed
-
- Yates JR, III, Griffin P, Hood L, Zhou J. Computer aided interpretation of low energy MS/MS mass spectra of peptides. Techniques in Protein Chemistry II. 1991;46:477–485.
-
- Owens KG. Application of Correlation Analysis Techniques to Mass Spectral Data. Applied Spectroscopy Reviews. 1992;27:1–49.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

