Group II intron lariat: Structural insights into the spliceosome
- PMID: 26121424
- PMCID: PMC4615233
- DOI: 10.1080/15476286.2015.1066956
Group II intron lariat: Structural insights into the spliceosome
Abstract
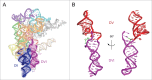
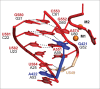
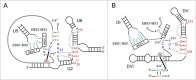
Group II introns are self-splicing catalytic RNAs found in bacteria and the organelles of fungi and plants. They are thought to share a common ancestor with the spliceosome, which catalyzes the removal of nuclear introns from pre-mRNAs in eukaryotes. Recent structural and biochemical evidence supports the hypothesis that the spliceosome has a catalytic RNA core homologous to that found in group II introns. The crystal structure of a eukaryotic group IIB intron was recently determined and reveals the architecture of a branched lariat RNA that is also formed by the spliceosome. Here we describe the active site components of this intron and propose a model for RNA splicing involving dynamic base triples in the catalytic triad. Based on this structure, we draw analogies to the U2/U6 snRNA pairing and RNA-protein interactions that form in the active site of the spliceosome.
Keywords: RNA splicing; branch point; group II intron; lariat; spliceosome.
Figures
Similar articles
-
Structural basis of pre-mRNA splicing.Science. 2015 Sep 11;349(6253):1191-8. doi: 10.1126/science.aac8159. Epub 2015 Aug 20. Science. 2015. PMID: 26292705
-
Crystal structures of a group II intron lariat primed for reverse splicing.Science. 2016 Dec 2;354(6316):aaf9258. doi: 10.1126/science.aaf9258. Science. 2016. PMID: 27934709
-
Cryo-EM structure of the spliceosome immediately after branching.Nature. 2016 Sep 8;537(7619):197-201. doi: 10.1038/nature19316. Epub 2016 Jul 26. Nature. 2016. PMID: 27459055 Free PMC article.
-
RNA Splicing by the Spliceosome.Annu Rev Biochem. 2020 Jun 20;89:359-388. doi: 10.1146/annurev-biochem-091719-064225. Epub 2019 Dec 3. Annu Rev Biochem. 2020. PMID: 31794245 Review.
-
The Spliceosome: A Protein-Directed Metalloribozyme.J Mol Biol. 2017 Aug 18;429(17):2640-2653. doi: 10.1016/j.jmb.2017.07.010. Epub 2017 Jul 19. J Mol Biol. 2017. PMID: 28733144 Review.
Cited by
-
SHAPE Profiling to Probe Group II Intron Conformational Dynamics During Splicing.Methods Mol Biol. 2021;2167:171-182. doi: 10.1007/978-1-0716-0716-9_10. Methods Mol Biol. 2021. PMID: 32712920 Free PMC article.
-
Understanding the mechanistic basis of non-coding RNA through molecular dynamics simulations.J Struct Biol. 2019 Jun 1;206(3):267-279. doi: 10.1016/j.jsb.2019.03.004. Epub 2019 Mar 15. J Struct Biol. 2019. PMID: 30880083 Free PMC article. Review.
-
Elimination of zero-repeat subunit in allergenic seed protein 13S globulin using the novel allele GlbNB2 in common buckwheat (Fagopyrum esculentum Moench).Food Chem (Oxf). 2024 Apr 22;8:100205. doi: 10.1016/j.fochms.2024.100205. eCollection 2024 Jul 30. Food Chem (Oxf). 2024. PMID: 38694165 Free PMC article.
-
Mobile Group II Introns as Ancestral Eukaryotic Elements.Trends Genet. 2017 Nov;33(11):773-783. doi: 10.1016/j.tig.2017.07.009. Epub 2017 Aug 14. Trends Genet. 2017. PMID: 28818345 Free PMC article. Review.
-
Categorizing 161 plant (streptophyte) mitochondrial group II introns into 29 families of related paralogues finds only limited links between intron mobility and intron-borne maturases.BMC Ecol Evol. 2023 Mar 13;23(1):5. doi: 10.1186/s12862-023-02108-y. BMC Ecol Evol. 2023. PMID: 36915058 Free PMC article.
References
-
- Lambowitz AM, Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol 2011; 3:a003616; PMID:20463000; http://dx.doi.org/10.1101/cshperspect.a003616 - DOI - PMC - PubMed
-
- Peebles CL, Perlman PS, Mecklenburg KL, Petrillo ML, Tabor JH, Jarrell KA, Cheng HL. A self-splicing RNA excises an intron lariat. Cell 1986; 44:213-23; PMID:3510741 - PubMed
-
- van der Veen R, Arnberg AC, van der Horst G, Bonen L, Tabak HF, Grivell LA. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell 1986; 44:225-34; PMID:2417726 - PubMed
-
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. RNA catalyses nuclear pre-mRNA splicing. Nature 2013; 503:229-34; PMID:24196718; http://dx.doi.org/10.1038/nature12734 - DOI - PMC - PubMed
-
- Valadkhan S, Mohammadi A, Wachtel C, Manley JL. Protein-free spliceosomal snRNAs catalyze a reaction that resembles the first step of splicing. RNA 2007; 13:2300-11; PMID:17940139; http://dx.doi.org/10.1261/rna.626207 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
