RNA BIOCHEMISTRY. Factor-dependent processivity in human eIF4A DEAD-box helicase
- PMID: 26113725
- PMCID: PMC4605566
- DOI: 10.1126/science.aaa5089
RNA BIOCHEMISTRY. Factor-dependent processivity in human eIF4A DEAD-box helicase
Abstract
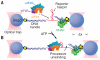
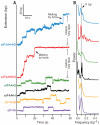
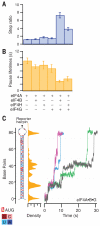
During eukaryotic translation initiation, the small ribosomal subunit, assisted by initiation factors, locates the messenger RNA start codon by scanning from the 5' cap. This process is powered by the eukaryotic initiation factor 4A (eIF4A), a DEAD-box helicase. eIF4A has been thought to unwind structures formed in the untranslated 5' region via a nonprocessive mechanism. Using a single-molecule assay, we found that eIF4A functions instead as an adenosine triphosphate-dependent processive helicase when complexed with two accessory proteins, eIF4G and eIF4B. Translocation occurred in discrete steps of 11 ± 2 base pairs, irrespective of the accessory factor combination. Our findings support a memory-less stepwise mechanism for translation initiation and suggest that similar factor-dependent processivity may be shared by other members of the DEAD-box helicase family.
Copyright © 2015, American Association for the Advancement of Science.
Figures
Similar articles
-
eIF4B and eIF4G jointly stimulate eIF4A ATPase and unwinding activities by modulation of the eIF4A conformational cycle.J Mol Biol. 2014 Jan 9;426(1):51-61. doi: 10.1016/j.jmb.2013.09.027. Epub 2013 Sep 27. J Mol Biol. 2014. PMID: 24080224
-
eIF4B stimulates eIF4A ATPase and unwinding activities by direct interaction through its 7-repeats region.RNA Biol. 2017 Jan 2;14(1):113-123. doi: 10.1080/15476286.2016.1259782. Epub 2016 Nov 18. RNA Biol. 2017. PMID: 27858515 Free PMC article.
-
eIF4B stimulates translation of long mRNAs with structured 5' UTRs and low closed-loop potential but weak dependence on eIF4G.Proc Natl Acad Sci U S A. 2016 Sep 20;113(38):10464-72. doi: 10.1073/pnas.1612398113. Epub 2016 Sep 6. Proc Natl Acad Sci U S A. 2016. PMID: 27601676 Free PMC article.
-
The DEAD-box helicase eIF4A: paradigm or the odd one out?RNA Biol. 2013 Jan;10(1):19-32. doi: 10.4161/rna.21966. Epub 2012 Sep 20. RNA Biol. 2013. PMID: 22995829 Free PMC article. Review.
-
eIF4A: the godfather of the DEAD box helicases.Prog Nucleic Acid Res Mol Biol. 2002;72:307-31. doi: 10.1016/s0079-6603(02)72073-4. Prog Nucleic Acid Res Mol Biol. 2002. PMID: 12206455 Review.
Cited by
-
The structure of a human translation initiation complex reveals two independent roles for the helicase eIF4A.Nat Struct Mol Biol. 2024 Mar;31(3):455-464. doi: 10.1038/s41594-023-01196-0. Epub 2024 Jan 29. Nat Struct Mol Biol. 2024. PMID: 38287194 Free PMC article.
-
The molecular basis of translation initiation and its regulation in eukaryotes.Nat Rev Mol Cell Biol. 2024 Mar;25(3):168-186. doi: 10.1038/s41580-023-00624-9. Epub 2023 Dec 5. Nat Rev Mol Cell Biol. 2024. PMID: 38052923 Review.
-
Control of Translation at the Initiation Phase During Glucose Starvation in Yeast.Int J Mol Sci. 2019 Aug 19;20(16):4043. doi: 10.3390/ijms20164043. Int J Mol Sci. 2019. PMID: 31430885 Free PMC article. Review.
-
Toward a Kinetic Understanding of Eukaryotic Translation.Cold Spring Harb Perspect Biol. 2019 Feb 1;11(2):a032706. doi: 10.1101/cshperspect.a032706. Cold Spring Harb Perspect Biol. 2019. PMID: 29959192 Free PMC article. Review.
-
Translational control of stem cell function.Nat Rev Mol Cell Biol. 2021 Oct;22(10):671-690. doi: 10.1038/s41580-021-00386-2. Epub 2021 Jul 16. Nat Rev Mol Cell Biol. 2021. PMID: 34272502 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

