The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation
- PMID: 26111795
- PMCID: PMC4500283
- DOI: 10.1073/pnas.1424220112
The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation
Abstract
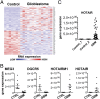
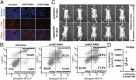
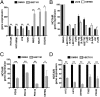
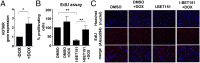
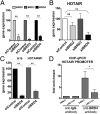
Bromodomain and extraterminal (BET) domain proteins have emerged as promising therapeutic targets in glioblastoma and many other cancers. Small molecule inhibitors of BET bromodomain proteins reduce expression of several oncogenes required for Glioblastoma Multiforme (GBM) progression. However, the mechanism through which BET protein inhibition reduces GBM growth is not completely understood. Long noncoding RNAs (lncRNAs) are important epigenetic regulators with critical roles in cancer initiation and malignant progression, but mechanistic insight into their expression and regulation by BET bromodomain inhibitors remains elusive. In this study, we used Helicos single molecule sequencing to comprehensively profile lncRNAs differentially expressed in GBM, and we identified a subset of GBM-specific lncRNAs whose expression is regulated by BET proteins. Treatment of GBM cells with the BET bromdomain inhibitor I-BET151 reduced levels of the tumor-promoting lncRNA HOX transcript antisense RNA (HOTAIR) and restored the expression of several other GBM down-regulated lncRNAs. Conversely, overexpression of HOTAIR in conjunction with I-BET151 treatment abrogates the antiproliferative activity of the BET bromodomain inhibitor. Moreover, chromatin immunoprecipitation analysis demonstrated binding of Bromodomain Containing 4 (BRD4) to the HOTAIR promoter, suggesting that BET proteins can directly regulate lncRNA expression. Our data unravel a previously unappreciated mechanism through which BET proteins control tumor growth of glioblastoma cells and suggest that modulation of lncRNA networks may, in part, mediate the antiproliferative effects of many epigenetic inhibitors currently in clinical trials for cancer and other diseases.
Keywords: BRD4; I-BET151; epigenetics; glioblastoma; long noncoding RNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
BET bromodomain proteins are required for glioblastoma cell proliferation.Epigenetics. 2014 Apr;9(4):611-20. doi: 10.4161/epi.27906. Epub 2014 Feb 19. Epigenetics. 2014. PMID: 24496381 Free PMC article.
-
The BET bromodomain inhibitor I-BET151 acts downstream of smoothened protein to abrogate the growth of hedgehog protein-driven cancers.J Biol Chem. 2014 Dec 19;289(51):35494-502. doi: 10.1074/jbc.M114.595348. Epub 2014 Oct 29. J Biol Chem. 2014. PMID: 25355313 Free PMC article.
-
Long non-coding RNA CASP5 promotes the malignant phenotypes of human glioblastoma multiforme.Biochem Biophys Res Commun. 2018 Jun 12;500(4):966-972. doi: 10.1016/j.bbrc.2018.04.217. Epub 2018 May 2. Biochem Biophys Res Commun. 2018. PMID: 29715460
-
Drug-induced modifications and modulations of microRNAs and long non-coding RNAs for future therapy against Glioblastoma Multiforme.Gene. 2020 Jan 10;723:144126. doi: 10.1016/j.gene.2019.144126. Epub 2019 Oct 4. Gene. 2020. PMID: 31589963 Review.
-
Natural flavonoids alleviate glioblastoma multiforme by regulating long non-coding RNA.Biomed Pharmacother. 2023 May;161:114477. doi: 10.1016/j.biopha.2023.114477. Epub 2023 Mar 15. Biomed Pharmacother. 2023. PMID: 36931030 Review.
Cited by
-
The Long Non-coding RNA HIF1A-AS2 Facilitates the Maintenance of Mesenchymal Glioblastoma Stem-like Cells in Hypoxic Niches.Cell Rep. 2016 Jun 14;15(11):2500-9. doi: 10.1016/j.celrep.2016.05.018. Epub 2016 Jun 2. Cell Rep. 2016. PMID: 27264189 Free PMC article.
-
LncRNAs associated with glioblastoma: From transcriptional noise to novel regulators with a promising role in therapeutics.Mol Ther Nucleic Acids. 2021 Apr 2;24:728-742. doi: 10.1016/j.omtn.2021.03.018. eCollection 2021 Jun 4. Mol Ther Nucleic Acids. 2021. PMID: 33996255 Free PMC article. Review.
-
Network-based modelling reveals cell-type enriched patterns of non-coding RNA regulation during human skeletal muscle remodelling.bioRxiv [Preprint]. 2024 Oct 9:2024.08.11.606848. doi: 10.1101/2024.08.11.606848. bioRxiv. 2024. Update in: NAR Mol Med. 2024 Oct 22;1(4):ugae016. doi: 10.1093/narmme/ugae016 PMID: 39416175 Free PMC article. Updated. Preprint.
-
Drug and disease signature integration identifies synergistic combinations in glioblastoma.Nat Commun. 2018 Dec 14;9(1):5315. doi: 10.1038/s41467-018-07659-z. Nat Commun. 2018. PMID: 30552330 Free PMC article.
-
The novel BET inhibitor UM-002 reduces glioblastoma cell proliferation and invasion.Sci Rep. 2021 Dec 3;11(1):23370. doi: 10.1038/s41598-021-02584-6. Sci Rep. 2021. PMID: 34862404 Free PMC article.
References
-
- Stupp R, et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. - PubMed
-
- Papavassiliou KA, Papavassiliou AG. Bromodomains: Pockets with therapeutic potential. Trends Mol Med. 2014;20(9):477–478. - PubMed
-
- Filippakopoulos P, Knapp S. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13(5):337–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

