Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage Trafficking
- PMID: 26107875
- PMCID: PMC4479461
- DOI: 10.1371/journal.ppat.1005004
Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage Trafficking
Abstract
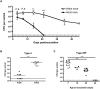
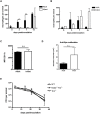
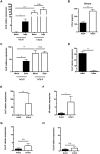
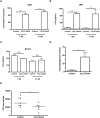
Infections are a common cause of infant mortality worldwide, especially due to Streptococcus pneumoniae. Colonization is the prerequisite to invasive pneumococcal disease, and is particularly frequent and prolonged in children, though the mechanisms underlying this susceptibility are unknown. We find that infant mice exhibit prolonged pneumococcal carriage, and are delayed in recruiting macrophages, the effector cells of clearance, into the nasopharyngeal lumen. This lack of macrophage recruitment is paralleled by a failure to upregulate chemokine (C-C) motif ligand 2 (Ccl2 or Mcp-1), a macrophage chemoattractant that is required in adult mice to promote clearance. Baseline expression of Ccl2 and the related chemokine Ccl7 is higher in the infant compared to the adult upper respiratory tract, and this effect requires the infant microbiota. These results demonstrate that signals governing macrophage recruitment are altered at baseline in infant mice, which prevents the development of appropriate innate cell infiltration in response to pneumococcal colonization, delaying clearance of pneumococcal carriage.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Impaired innate and adaptive immunity to Streptococcus pneumoniae and its effect on colonization in an infant mouse model.Infect Immun. 2009 Apr;77(4):1613-22. doi: 10.1128/IAI.00871-08. Epub 2009 Jan 21. Infect Immun. 2009. PMID: 19168741 Free PMC article.
-
Macrolides Promote CCL2-Mediated Macrophage Recruitment and Clearance of Nasopharyngeal Pneumococcal Colonization in Mice.J Infect Dis. 2015 Oct 1;212(7):1150-9. doi: 10.1093/infdis/jiv157. Epub 2015 Mar 12. J Infect Dis. 2015. PMID: 25767216
-
Macrophage migration inhibitory factor promotes clearance of pneumococcal colonization.J Immunol. 2014 Jul 15;193(2):764-72. doi: 10.4049/jimmunol.1400133. Epub 2014 Jun 13. J Immunol. 2014. PMID: 24928996 Free PMC article.
-
Nasopharyngeal Bacterial Carriage in the Conjugate Vaccine Era with a Focus on Pneumococci.J Immunol Res. 2015;2015:394368. doi: 10.1155/2015/394368. Epub 2015 Aug 16. J Immunol Res. 2015. PMID: 26351646 Free PMC article. Review.
-
The fundamental link between pneumococcal carriage and disease.Expert Rev Vaccines. 2012 Jul;11(7):841-55. doi: 10.1586/erv.12.53. Expert Rev Vaccines. 2012. PMID: 22913260 Review.
Cited by
-
Composition and immunological significance of the upper respiratory tract microbiota.FEBS Lett. 2016 Nov;590(21):3705-3720. doi: 10.1002/1873-3468.12455. Epub 2016 Nov 1. FEBS Lett. 2016. PMID: 27730630 Free PMC article. Review.
-
Purified Streptococcus pneumoniae Endopeptidase O (PepO) Enhances Particle Uptake by Macrophages in a Toll-Like Receptor 2- and miR-155-Dependent Manner.Infect Immun. 2017 Mar 23;85(4):e01012-16. doi: 10.1128/IAI.01012-16. Print 2017 Apr. Infect Immun. 2017. PMID: 28193634 Free PMC article.
-
Host-to-Host Transmission of Streptococcus pneumoniae Is Driven by Its Inflammatory Toxin, Pneumolysin.Cell Host Microbe. 2017 Jan 11;21(1):73-83. doi: 10.1016/j.chom.2016.12.005. Cell Host Microbe. 2017. PMID: 28081446 Free PMC article.
-
Fatal Pertussis in the Neonatal Mouse Model Is Associated with Pertussis Toxin-Mediated Pathology beyond the Airways.Infect Immun. 2017 Oct 18;85(11):e00355-17. doi: 10.1128/IAI.00355-17. Print 2017 Nov. Infect Immun. 2017. PMID: 28784932 Free PMC article.
-
Pneumococcal quorum sensing drives an asymmetric owner-intruder competitive strategy during carriage via the competence regulon.Nat Microbiol. 2019 Jan;4(1):198-208. doi: 10.1038/s41564-018-0314-4. Epub 2018 Dec 10. Nat Microbiol. 2019. PMID: 30546100 Free PMC article.
References
-
- Gray BM, Converse GM, Dillon HC (1980) Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J Infect Dis 142: 923–933. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

