c-di-GMP and its Effects on Biofilm Formation and Dispersion: a Pseudomonas Aeruginosa Review
- PMID: 26104694
- PMCID: PMC4498269
- DOI: 10.1128/microbiolspec.MB-0003-2014
c-di-GMP and its Effects on Biofilm Formation and Dispersion: a Pseudomonas Aeruginosa Review
Abstract
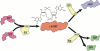
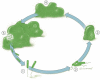
Since its initial discovery as an allosteric factor regulating cellulose biosynthesis in Gluconacetobacter xylinus, the list of functional outputs regulated by c-di-GMP has grown. We have focused this article on one of these c-di-GMP-regulated processes, namely, biofilm formation in the organism Pseudomonas aeruginosa. The majority of diguanylate cyclases and phosphodiesterases encoded in the P. aeruginosa genome still remain uncharacterized; thus, there is still a great deal to be learned about the link between c-di-GMP and biofilm formation in this microbe. In particular, while a number of c-di-GMP metabolizing enzymes have been identified that participate in reversible and irreversible attachment and biofilm maturation, there is a still a significant knowledge gap regarding the c-di-GMP output systems in this organism. Even for the well-characterized Pel system, where c-di-GMP-mediated transcriptional regulation is now well documented, how binding of c-di-GMP by PelD stimulates Pel production is not understood in any detail. Similarly, c-di-GMP-mediated control of swimming, swarming and twitching also remains to be elucidated. Thus, despite terrific advances in our understanding of P. aeruginosa biofilm formation and the role of c-di-GMP in this process since the last version of this book (indeed there was no chapter on c-di-GMP!) there is still much to learn.
Figures
Similar articles
-
Pseudomonas aeruginosa Uses c-di-GMP Phosphodiesterases RmcA and MorA To Regulate Biofilm Maintenance.mBio. 2021 Feb 2;12(1):e03384-20. doi: 10.1128/mBio.03384-20. mBio. 2021. PMID: 33531388 Free PMC article.
-
Diguanylate Cyclases and Phosphodiesterases Required for Basal-Level c-di-GMP in Pseudomonas aeruginosa as Revealed by Systematic Phylogenetic and Transcriptomic Analyses.Appl Environ Microbiol. 2019 Oct 16;85(21):e01194-19. doi: 10.1128/AEM.01194-19. Print 2019 Nov 1. Appl Environ Microbiol. 2019. PMID: 31444209 Free PMC article.
-
Role of Cyclic Di-GMP and Exopolysaccharide in Type IV Pilus Dynamics.J Bacteriol. 2017 Mar 28;199(8):e00859-16. doi: 10.1128/JB.00859-16. Print 2017 Apr 15. J Bacteriol. 2017. PMID: 28167523 Free PMC article.
-
Biofilms and Cyclic di-GMP (c-di-GMP) Signaling: Lessons from Pseudomonas aeruginosa and Other Bacteria.J Biol Chem. 2016 Jun 10;291(24):12547-12555. doi: 10.1074/jbc.R115.711507. Epub 2016 Apr 21. J Biol Chem. 2016. PMID: 27129226 Free PMC article. Review.
-
Molecular and structural facets of c-di-GMP signalling associated with biofilm formation in Pseudomonas aeruginosa.Mol Aspects Med. 2021 Oct;81:101001. doi: 10.1016/j.mam.2021.101001. Epub 2021 Jul 24. Mol Aspects Med. 2021. PMID: 34311995 Review.
Cited by
-
The Nitrite Transporter Facilitates Biofilm Formation via Suppression of Nitrite Reductase and Is a New Antibiofilm Target in Pseudomonas aeruginosa.mBio. 2020 Jul 7;11(4):e00878-20. doi: 10.1128/mBio.00878-20. mBio. 2020. PMID: 32636243 Free PMC article.
-
PP4397/FlgZ provides the link between PP2258 c-di-GMP signalling and altered motility in Pseudomonas putida.Sci Rep. 2018 Aug 15;8(1):12205. doi: 10.1038/s41598-018-29785-w. Sci Rep. 2018. PMID: 30111852 Free PMC article.
-
PqsA Promotes Pyoverdine Production via Biofilm Formation.Pathogens. 2017 Dec 25;7(1):3. doi: 10.3390/pathogens7010003. Pathogens. 2017. PMID: 29295589 Free PMC article.
-
Biofilm development of an opportunistic model bacterium analysed at high spatiotemporal resolution in the framework of a precise flow cell.NPJ Biofilms Microbiomes. 2016 Oct 19;2:16023. doi: 10.1038/npjbiofilms.2016.23. eCollection 2016. NPJ Biofilms Microbiomes. 2016. PMID: 28721252 Free PMC article.
-
The effect of Cyclic-di-GMP on biofilm formation by Pseudomonas aeruginosa in a novel empyema model.Ann Transl Med. 2020 Sep;8(18):1146. doi: 10.21037/atm-20-6022. Ann Transl Med. 2020. PMID: 33240995 Free PMC article.
References
-
- Sutherland IW. The biofilm matrix--an immobilized but dynamic microbial environment. Trends Microbiol. 2001;9:222–227. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

