Endoplasmic Reticulum Tubule Protein Reticulon 4 Associates with the Legionella pneumophila Vacuole and with Translocated Substrate Ceg9
- PMID: 26099580
- PMCID: PMC4534651
- DOI: 10.1128/IAI.00507-15
Endoplasmic Reticulum Tubule Protein Reticulon 4 Associates with the Legionella pneumophila Vacuole and with Translocated Substrate Ceg9
Abstract
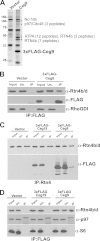
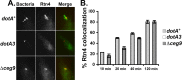
Intracellular growth of Legionella pneumophila occurs in a replication vacuole constructed by host proteins that regulate vesicular traffic from the host endoplasmic reticulum (ER). This process is promoted by a combination of approximately 300 Icm/Dot translocated substrates (IDTS). One of these proteins, Ceg9, was previously identified in a screen for L. pneumophila IDTS that manipulate secretory traffic when overexpressed in yeast. Using ectopic expression of Ceg9 in mammalian cells, we demonstrate that Ceg9 interacts with isoforms of host reticulon 4 (Rtn4), a protein that regulates ER tubule formation. Binding occurs under conditions that prevent association with other known reticulon binding proteins, arguing that Ceg9 binding is stable. A tripartite complex was demonstrated among Rtn4, Ceg9, and atlastin 1, a previously characterized reticulon interacting partner. The binding of Ceg9 to Rtn4 was not due to bridging by atlastin 1 but resulted from the two interacting partners binding independently to reticulon. When Ceg9 is ectopically expressed in mammalian cells, it shows a localization pattern that is indistinguishable from that of Rtn4, perhaps due to interactions between and similar structural features of the two proteins. Consistent with Rtn4 playing a role in the formation of the Legionella-containing vacuole, it was recruited to almost 50% of the vacuoles within 20 min postinfection. Our studies suggest that L. pneumophila proteins interact with ER tubules at an early stage of replication vacuole formation.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Legionella hijacks the host Golgi-to-ER retrograde pathway for the association of Legionella-containing vacuole with the ER.PLoS Pathog. 2021 Mar 24;17(3):e1009437. doi: 10.1371/journal.ppat.1009437. eCollection 2021 Mar. PLoS Pathog. 2021. PMID: 33760868 Free PMC article.
-
A Single Legionella Effector Catalyzes a Multistep Ubiquitination Pathway to Rearrange Tubular Endoplasmic Reticulum for Replication.Cell Host Microbe. 2017 Feb 8;21(2):169-181. doi: 10.1016/j.chom.2016.12.007. Epub 2016 Dec 29. Cell Host Microbe. 2017. PMID: 28041930 Free PMC article.
-
Autophagy Evasion and Endoplasmic Reticulum Subversion: The Yin and Yang of Legionella Intracellular Infection.Annu Rev Microbiol. 2016 Sep 8;70:413-33. doi: 10.1146/annurev-micro-102215-095557. Annu Rev Microbiol. 2016. PMID: 27607556 Review.
-
The Legionella effector RavD binds phosphatidylinositol-3-phosphate and helps suppress endolysosomal maturation of the Legionella-containing vacuole.J Biol Chem. 2019 Apr 19;294(16):6405-6415. doi: 10.1074/jbc.RA118.007086. Epub 2019 Feb 7. J Biol Chem. 2019. PMID: 30733336 Free PMC article.
-
The road less traveled: transport of Legionella to the endoplasmic reticulum.J Cell Biol. 2002 Aug 5;158(3):415-9. doi: 10.1083/jcb.200205011. Epub 2002 Jul 29. J Cell Biol. 2002. PMID: 12147677 Free PMC article. Review.
Cited by
-
Evolution and Adaptation of Legionella pneumophila to Manipulate the Ubiquitination Machinery of Its Amoebae and Mammalian Hosts.Biomolecules. 2021 Jan 15;11(1):112. doi: 10.3390/biom11010112. Biomolecules. 2021. PMID: 33467718 Free PMC article. Review.
-
Sde proteins coordinate ubiquitin utilization and phosphoribosylation to establish and maintain the Legionella replication vacuole.Nat Commun. 2024 Aug 30;15(1):7479. doi: 10.1038/s41467-024-51272-2. Nat Commun. 2024. PMID: 39214970 Free PMC article.
-
Type II Secretion Is Necessary for Optimal Association of the Legionella-Containing Vacuole with Macrophage Rab1B but Enhances Intracellular Replication Mainly by Rab1B-Independent Mechanisms.Infect Immun. 2016 Nov 18;84(12):3313-3327. doi: 10.1128/IAI.00750-16. Print 2016 Dec. Infect Immun. 2016. PMID: 27600508 Free PMC article.
-
Hostile Takeover: Hijacking of Endoplasmic Reticulum Function by T4SS and T3SS Effectors Creates a Niche for Intracellular Pathogens.Microbiol Spectr. 2019 May;7(3):10.1128/microbiolspec.psib-0027-2019. doi: 10.1128/microbiolspec.PSIB-0027-2019. Microbiol Spectr. 2019. PMID: 31198132 Free PMC article.
-
Amoebae as training grounds for microbial pathogens.mBio. 2024 Aug 14;15(8):e0082724. doi: 10.1128/mbio.00827-24. Epub 2024 Jul 8. mBio. 2024. PMID: 38975782 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

