HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen
- PMID: 26089355
- PMCID: PMC4669217
- DOI: 10.1126/science.aac5894
HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen
Abstract
A major goal of HIV-1 vaccine research is the design of immunogens capable of inducing broadly neutralizing antibodies (bnAbs) that bind to the viral envelope glycoprotein (Env). Poor binding of Env to unmutated precursors of bnAbs, including those of the VRC01 class, appears to be a major problem for bnAb induction. We engineered an immunogen that binds to VRC01-class bnAb precursors and immunized knock-in mice expressing germline-reverted VRC01 heavy chains. Induced antibodies showed characteristics of VRC01-class bnAbs, including a short CDRL3 (light-chain complementarity-determining region 3) and mutations that favored binding to near-native HIV-1 gp120 constructs. In contrast, native-like immunogens failed to activate VRC01-class precursors. The results suggest that rational epitope design can prime rare B cell precursors for affinity maturation to desired targets.
Copyright © 2015, American Association for the Advancement of Science.
Figures
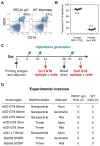
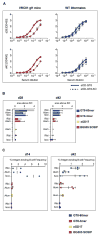
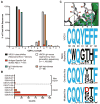
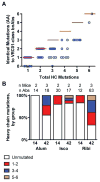

Comment in
-
HIV. The modern era of HIV-1 vaccine development.Science. 2015 Jul 10;349(6244):139-40. doi: 10.1126/science.aac7800. Science. 2015. PMID: 26160931 No abstract available.
Similar articles
-
Short CDRL1 in intermediate VRC01-like mAbs is not sufficient to overcome key glycan barriers on HIV-1 Env.J Virol. 2024 Oct 22;98(10):e0074424. doi: 10.1128/jvi.00744-24. Epub 2024 Sep 6. J Virol. 2024. PMID: 39240111 Free PMC article.
-
HIV-1 envelope glycan modifications that permit neutralization by germline-reverted VRC01-class broadly neutralizing antibodies.PLoS Pathog. 2018 Nov 5;14(11):e1007431. doi: 10.1371/journal.ppat.1007431. eCollection 2018 Nov. PLoS Pathog. 2018. PMID: 30395637 Free PMC article.
-
Glycan Masking Focuses Immune Responses to the HIV-1 CD4-Binding Site and Enhances Elicitation of VRC01-Class Precursor Antibodies.Immunity. 2018 Aug 21;49(2):301-311.e5. doi: 10.1016/j.immuni.2018.07.005. Epub 2018 Jul 31. Immunity. 2018. PMID: 30076101 Free PMC article.
-
HIV Broadly Neutralizing Antibodies: VRC01 and Beyond.Adv Exp Med Biol. 2018;1075:53-72. doi: 10.1007/978-981-13-0484-2_3. Adv Exp Med Biol. 2018. PMID: 30030789 Review.
-
Targeting broadly neutralizing antibody precursors: a naïve approach to vaccine design.Curr Opin HIV AIDS. 2019 Jul;14(4):294-301. doi: 10.1097/COH.0000000000000548. Curr Opin HIV AIDS. 2019. PMID: 30946041 Review.
Cited by
-
Design of immunogens to elicit broadly neutralizing antibodies against HIV targeting the CD4 binding site.Proc Natl Acad Sci U S A. 2021 Mar 2;118(9):e2018338118. doi: 10.1073/pnas.2018338118. Proc Natl Acad Sci U S A. 2021. PMID: 33637649 Free PMC article.
-
HIV-1 VRC01 Germline-Targeting Immunogens Select Distinct Epitope-Specific B Cell Receptors.Immunity. 2020 Oct 13;53(4):840-851.e6. doi: 10.1016/j.immuni.2020.09.007. Immunity. 2020. PMID: 33053332 Free PMC article.
-
Influence of glycosylation on the immunogenicity and antigenicity of viral immunogens.Biotechnol Adv. 2024 Jan-Feb;70:108283. doi: 10.1016/j.biotechadv.2023.108283. Epub 2023 Nov 14. Biotechnol Adv. 2024. PMID: 37972669 Free PMC article. Review.
-
Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G.Cell. 2016 May 5;165(4):813-26. doi: 10.1016/j.cell.2016.04.010. Epub 2016 Apr 21. Cell. 2016. PMID: 27114034 Free PMC article.
-
Development of broadly neutralizing antibodies in HIV-1 infected elite neutralizers.Retrovirology. 2018 Sep 5;15(1):61. doi: 10.1186/s12977-018-0443-0. Retrovirology. 2018. PMID: 30185183 Free PMC article. Review.
References
-
- Gauduin MC, et al. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nature medicine. 1997;3:1389–1393. - PubMed
-
- Mascola JR, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nature medicine. 2000;6:207–210. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

